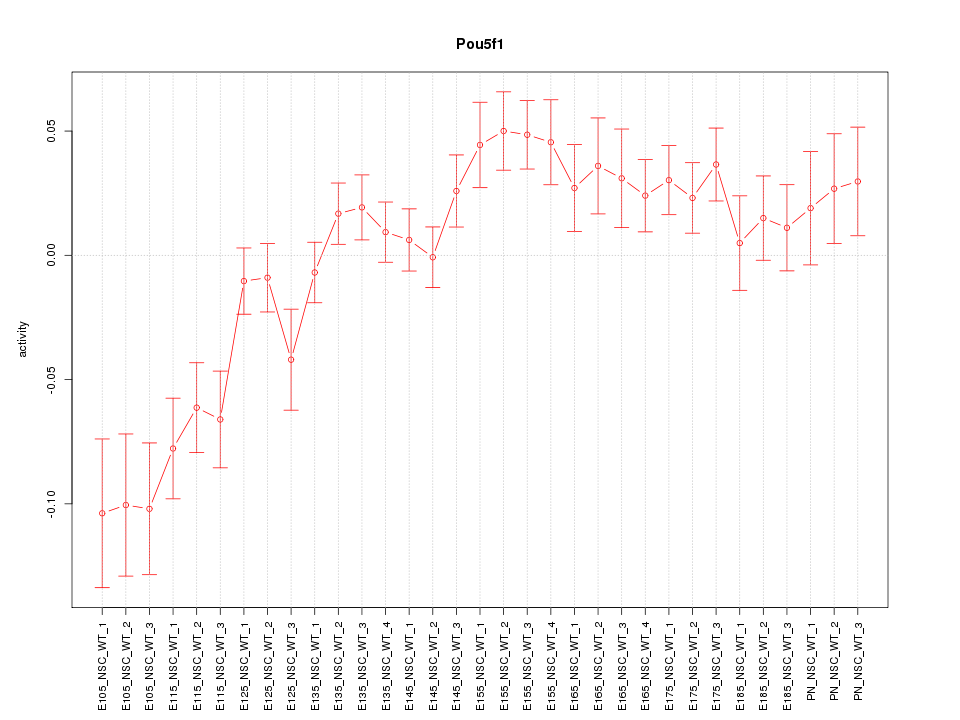
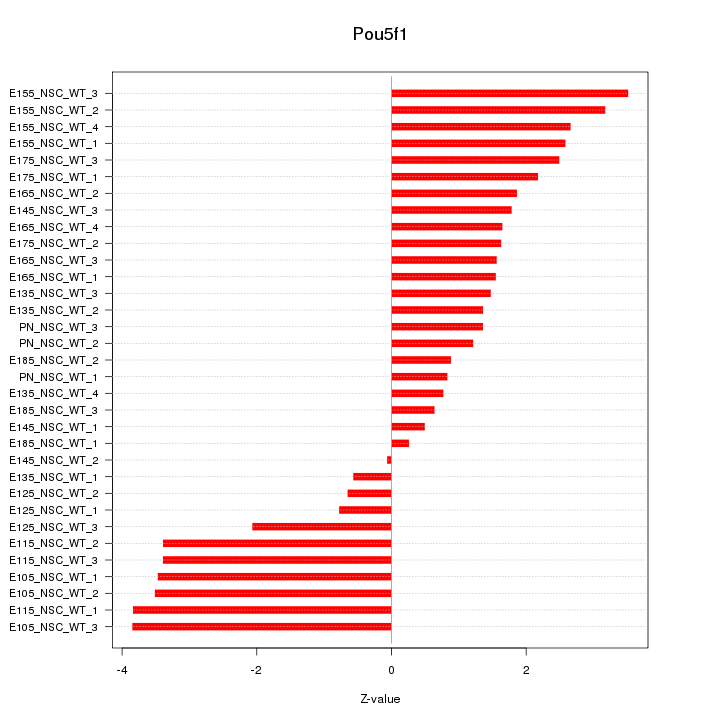
Motif ID: Pou5f1
Z-value: 2.179

Transcription factors associated with Pou5f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou5f1 | ENSMUSG00000024406.10 | Pou5f1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 5.2 | 20.9 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 4.9 | 24.6 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 4.6 | 32.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 4.5 | 18.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 4.3 | 42.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 3.2 | 44.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 2.7 | 8.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 2.5 | 10.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 2.5 | 7.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 2.3 | 11.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 2.2 | 8.9 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 2.2 | 6.5 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 2.1 | 6.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 1.8 | 12.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 11.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.6 | 4.8 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.6 | 9.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 1.6 | 4.7 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.6 | 23.3 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 1.5 | 7.7 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 1.5 | 6.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.4 | 47.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 1.4 | 2.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 1.4 | 9.5 | GO:0071361 | cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 1.3 | 27.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.2 | 3.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 1.2 | 14.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 1.1 | 4.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.1 | 4.5 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.1 | 14.9 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 1.0 | 11.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 1.0 | 3.9 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 1.0 | 5.7 | GO:1903056 | melanocyte migration(GO:0097324) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.9 | 4.4 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.9 | 5.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.9 | 2.6 | GO:0019043 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.8 | 3.3 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.8 | 2.5 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.8 | 5.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.8 | 4.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.7 | 77.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.7 | 26.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.7 | 22.4 | GO:0007616 | long-term memory(GO:0007616) |
| 0.7 | 2.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.7 | 5.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.6 | 18.7 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.6 | 1.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.5 | 1.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 3.7 | GO:0001696 | gastric acid secretion(GO:0001696) oxalate transport(GO:0019532) |
| 0.5 | 4.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.5 | 2.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.5 | 3.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.5 | 3.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 4.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.5 | 6.9 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.4 | 3.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 7.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.4 | 2.0 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.4 | 3.9 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.4 | 28.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 1.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.3 | 10.7 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.3 | 2.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.3 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.3 | 3.6 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.3 | 4.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 0.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 8.7 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.3 | 3.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 1.5 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 3.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 0.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 2.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 6.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 0.6 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.2 | 1.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 0.8 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 4.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 4.4 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.2 | 1.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 1.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.2 | 28.1 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.2 | 1.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 1.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 3.0 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.2 | 5.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 3.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 7.3 | GO:0060411 | cardiac septum morphogenesis(GO:0060411) |
| 0.1 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 4.3 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 5.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.8 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 6.5 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 4.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 2.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 2.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 6.3 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 1.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.5 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 3.9 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 7.9 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 2.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.3 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 2.6 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 2.8 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 1.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.4 | GO:0044308 | axonal spine(GO:0044308) |
| 2.6 | 18.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.7 | 32.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.4 | 23.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 1.3 | 20.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.2 | 4.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.9 | 9.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.9 | 11.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.8 | 10.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.8 | 7.6 | GO:0000805 | X chromosome(GO:0000805) |
| 0.7 | 23.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.7 | 14.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.7 | 2.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.6 | 41.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.6 | 23.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.5 | 25.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 2.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 3.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 5.9 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.4 | 3.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 1.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 5.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 1.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 1.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 4.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 9.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 11.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 8.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 0.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 1.0 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 71.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 2.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 10.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 9.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 9.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 3.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 8.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 16.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 4.9 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 3.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 23.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 6.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 3.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.3 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 5.7 | GO:0005768 | endosome(GO:0005768) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.6 | 42.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 7.7 | 23.1 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 6.2 | 43.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 3.8 | 11.3 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 3.5 | 20.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 3.2 | 9.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 2.3 | 32.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 2.0 | 24.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.8 | 18.1 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 1.8 | 8.9 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.6 | 6.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.4 | 9.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.3 | 6.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.2 | 3.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.2 | 3.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.2 | 5.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.1 | 14.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.1 | 3.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.0 | 4.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.9 | 4.4 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.8 | 3.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.7 | 5.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.7 | 2.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 2.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.6 | 4.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.6 | 20.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 4.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.5 | 5.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.5 | 3.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 5.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 3.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.4 | 8.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 4.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.4 | 3.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 4.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 31.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.3 | 3.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 2.4 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 5.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.3 | 10.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 12.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 6.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 11.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 1.1 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 2.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 10.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 3.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 1.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 2.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 3.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 6.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 2.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 5.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.3 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.1 | 4.9 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 9.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 0.6 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 2.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 6.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 11.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 6.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 4.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 5.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 4.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 2.6 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 14.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 4.8 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 1.5 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 2.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |