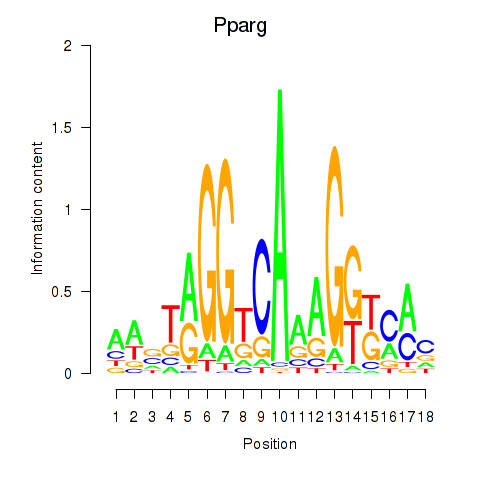
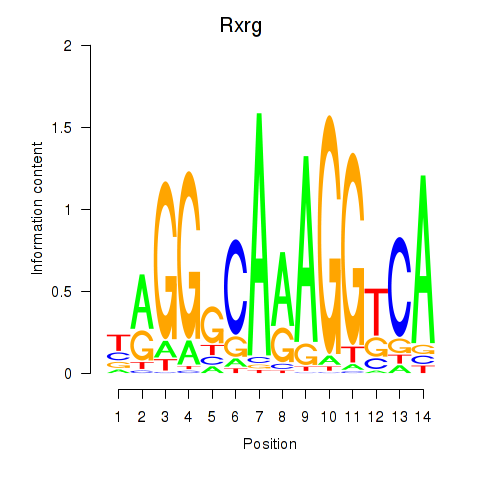
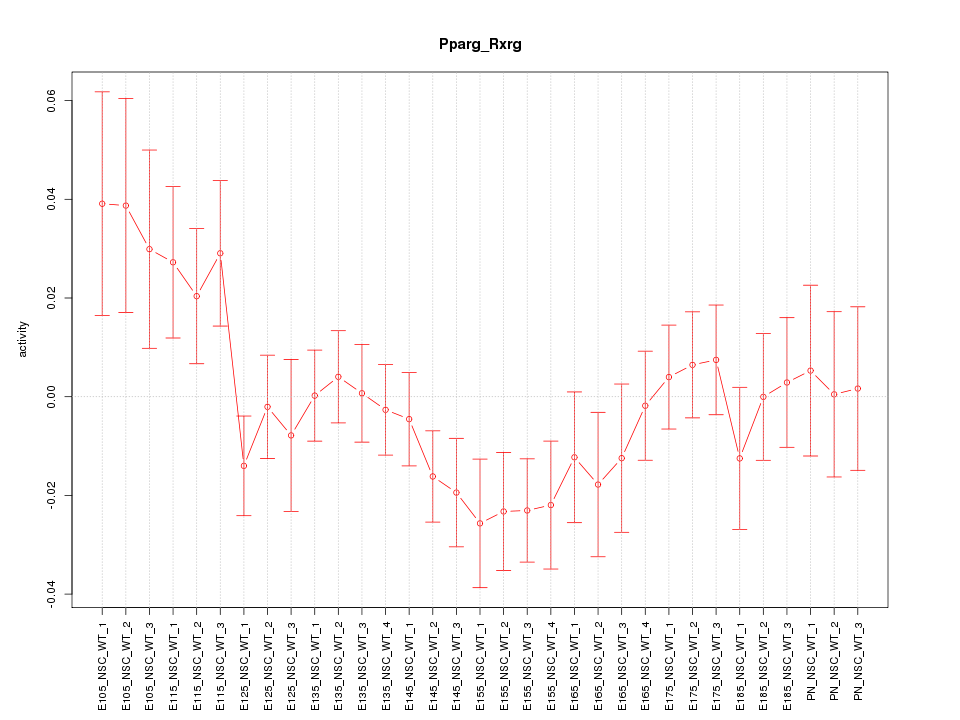
| 2.0 |
10.0 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 1.5 |
4.6 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.4 |
4.2 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 1.3 |
6.5 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.3 |
3.8 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.2 |
3.7 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 1.2 |
3.5 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 1.1 |
6.6 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 1.0 |
5.2 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 1.0 |
2.9 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.9 |
6.5 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.7 |
7.1 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.7 |
4.9 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.7 |
2.1 |
GO:0046416 |
D-amino acid catabolic process(GO:0019478) D-amino acid metabolic process(GO:0046416) |
| 0.7 |
4.0 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.7 |
2.0 |
GO:0060129 |
thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.7 |
2.0 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.7 |
2.0 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.6 |
2.5 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.6 |
2.5 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.6 |
4.3 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.6 |
1.9 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.6 |
1.8 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.6 |
2.4 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.6 |
7.5 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.6 |
2.9 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.5 |
1.6 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.5 |
3.1 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 |
3.6 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.5 |
1.6 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.5 |
1.5 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.5 |
2.0 |
GO:1902302 |
regulation of potassium ion export(GO:1902302) |
| 0.5 |
2.5 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.5 |
1.5 |
GO:1900211 |
positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) |
| 0.5 |
2.4 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.5 |
3.4 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.5 |
1.4 |
GO:1904192 |
cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.5 |
1.9 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.5 |
0.5 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.4 |
2.2 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 |
0.4 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.4 |
1.7 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 |
3.0 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.4 |
3.2 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 |
3.1 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.4 |
0.8 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.4 |
0.8 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 |
1.1 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.4 |
1.8 |
GO:0060334 |
regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.3 |
1.4 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.3 |
1.7 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.3 |
1.0 |
GO:0060084 |
synaptic transmission involved in micturition(GO:0060084) |
| 0.3 |
1.0 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 |
0.6 |
GO:0035771 |
interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.3 |
0.9 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.3 |
1.5 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.3 |
1.2 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 |
3.8 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 |
0.9 |
GO:0001803 |
type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 |
0.9 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.3 |
3.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.3 |
2.2 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.3 |
0.8 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.3 |
1.7 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 |
0.5 |
GO:0006145 |
purine nucleobase catabolic process(GO:0006145) |
| 0.3 |
0.8 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.3 |
1.9 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.3 |
0.8 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.3 |
1.0 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.3 |
0.8 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.2 |
5.2 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 |
1.0 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.2 |
1.0 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 |
1.2 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 |
1.7 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 |
1.0 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 |
0.7 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 |
0.9 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.2 |
0.7 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.2 |
1.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.2 |
2.3 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 |
8.2 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 |
0.7 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) retinal metabolic process(GO:0042574) |
| 0.2 |
1.7 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 |
0.6 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 |
1.5 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.2 |
1.1 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.2 |
0.4 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.2 |
1.5 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.2 |
1.2 |
GO:0060290 |
transdifferentiation(GO:0060290) |
| 0.2 |
1.4 |
GO:0033567 |
DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.2 |
1.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.2 |
0.6 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.2 |
0.4 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 |
0.8 |
GO:0046655 |
folic acid metabolic process(GO:0046655) pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.2 |
0.4 |
GO:0006987 |
activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.2 |
1.7 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.2 |
0.6 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.2 |
1.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 |
1.7 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.2 |
1.0 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.2 |
0.8 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 |
0.8 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 |
0.9 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 |
1.5 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.2 |
0.9 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.2 |
0.9 |
GO:0032055 |
negative regulation of translation in response to stress(GO:0032055) |
| 0.2 |
2.6 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.2 |
1.7 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 |
0.4 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.2 |
0.9 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.2 |
0.5 |
GO:0070120 |
ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.2 |
1.8 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 |
0.5 |
GO:1903596 |
regulation of gap junction assembly(GO:1903596) |
| 0.2 |
0.5 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.2 |
0.9 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.2 |
1.6 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 |
0.9 |
GO:0060279 |
positive regulation of ovulation(GO:0060279) |
| 0.2 |
0.9 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.2 |
0.5 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 |
2.2 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 |
1.5 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 |
0.7 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 |
0.8 |
GO:0046668 |
regulation of retinal cell programmed cell death(GO:0046668) |
| 0.2 |
0.6 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 |
0.5 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 |
1.9 |
GO:0051156 |
glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 |
1.6 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 |
0.3 |
GO:0002730 |
regulation of dendritic cell cytokine production(GO:0002730) |
| 0.2 |
0.6 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.2 |
0.5 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.2 |
0.9 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.2 |
0.5 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 |
1.5 |
GO:0032306 |
regulation of prostaglandin secretion(GO:0032306) |
| 0.2 |
0.5 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 |
1.0 |
GO:0032725 |
positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.1 |
1.0 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
0.4 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.4 |
GO:0018008 |
N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.1 |
2.8 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.4 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
2.2 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
1.3 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
0.6 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
0.4 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 |
2.0 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.1 |
3.2 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.1 |
3.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.6 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
3.8 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.1 |
2.5 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.1 |
0.9 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.1 |
0.4 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 |
0.5 |
GO:2000195 |
negative regulation of female gonad development(GO:2000195) |
| 0.1 |
0.6 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 |
0.4 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.6 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
0.8 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
1.6 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
0.4 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.1 |
0.7 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
1.1 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
1.7 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.1 |
0.6 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 |
0.4 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 |
0.2 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 |
0.5 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) negative regulation of t-circle formation(GO:1904430) |
| 0.1 |
0.6 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 |
0.3 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.1 |
0.8 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.1 |
1.3 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.9 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
0.4 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 |
0.4 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 |
1.1 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
0.4 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.1 |
0.6 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.1 |
3.5 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 |
2.9 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
1.9 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.1 |
0.5 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.1 |
0.7 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 |
2.1 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.1 |
0.7 |
GO:0061732 |
mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 |
0.2 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
3.2 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.1 |
6.2 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.1 |
0.6 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.1 |
0.4 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 |
0.2 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 |
0.4 |
GO:0018202 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.1 |
0.3 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 |
1.9 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 |
0.4 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
0.2 |
GO:0001798 |
type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type II hypersensitivity(GO:0002445) positive regulation of hypersensitivity(GO:0002885) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) type I hypersensitivity(GO:0016068) |
| 0.1 |
2.6 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.1 |
3.6 |
GO:0007588 |
excretion(GO:0007588) |
| 0.1 |
0.5 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
3.3 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.1 |
0.3 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.1 |
0.3 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.1 |
0.5 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
0.5 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.1 |
0.2 |
GO:0048710 |
regulation of astrocyte differentiation(GO:0048710) positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 |
0.3 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 |
1.4 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 |
0.1 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 |
1.4 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.1 |
1.2 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.1 |
1.2 |
GO:0043567 |
positive regulation of activated T cell proliferation(GO:0042104) regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 |
1.1 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 |
0.3 |
GO:0090110 |
cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
2.0 |
GO:0014009 |
glial cell proliferation(GO:0014009) |
| 0.1 |
1.0 |
GO:0001773 |
myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.8 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
1.4 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
0.9 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.6 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.1 |
0.8 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 |
1.3 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.1 |
0.3 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 |
0.1 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.1 |
0.5 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.2 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
1.1 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.1 |
0.9 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 |
0.6 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
1.2 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
0.5 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.9 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.1 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 |
0.3 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
1.4 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 |
0.4 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.1 |
0.9 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
0.7 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
0.5 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.5 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 |
0.1 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.1 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 |
0.3 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.1 |
1.4 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
0.4 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.4 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
3.0 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 |
0.7 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.1 |
0.2 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
2.9 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.1 |
1.6 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 |
0.2 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.4 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 |
0.8 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.1 |
0.8 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 |
0.7 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.9 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.1 |
0.4 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.7 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
0.5 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.1 |
3.8 |
GO:0008203 |
cholesterol metabolic process(GO:0008203) |
| 0.1 |
0.5 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 |
0.3 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 |
6.7 |
GO:0071229 |
cellular response to acid chemical(GO:0071229) |
| 0.1 |
0.2 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.1 |
1.4 |
GO:0042304 |
regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 |
0.3 |
GO:0043589 |
skin morphogenesis(GO:0043589) negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 |
0.3 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 |
0.6 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 |
1.2 |
GO:0072673 |
lamellipodium morphogenesis(GO:0072673) |
| 0.1 |
0.2 |
GO:0006788 |
heme oxidation(GO:0006788) |
| 0.1 |
0.6 |
GO:2000810 |
regulation of bicellular tight junction assembly(GO:2000810) |
| 0.1 |
1.9 |
GO:0097006 |
regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.1 |
0.4 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.1 |
1.3 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 |
0.5 |
GO:0006664 |
glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 |
0.5 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.9 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.3 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
2.1 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
1.7 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
1.1 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.2 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.1 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.0 |
0.4 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.3 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 |
0.0 |
GO:0010829 |
negative regulation of glucose transport(GO:0010829) |
| 0.0 |
0.6 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.2 |
GO:0097300 |
necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 |
0.1 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.3 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.3 |
GO:0030728 |
ovulation(GO:0030728) |
| 0.0 |
0.2 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
1.0 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.3 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
1.0 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 |
0.1 |
GO:0046016 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 |
0.5 |
GO:0006693 |
prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 |
0.3 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
4.8 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.9 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
1.0 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.5 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
1.1 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.3 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.5 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.9 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.0 |
GO:0060264 |
respiratory burst involved in inflammatory response(GO:0002536) lipopolysaccharide transport(GO:0015920) regulation of respiratory burst involved in inflammatory response(GO:0060264) positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 |
0.5 |
GO:0051043 |
regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 |
0.2 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.0 |
0.2 |
GO:0051084 |
'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 |
0.5 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.8 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.4 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.0 |
1.2 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.5 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.3 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.5 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.0 |
0.4 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
1.3 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
1.8 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.6 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
1.0 |
GO:0030166 |
proteoglycan biosynthetic process(GO:0030166) |
| 0.0 |
0.1 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.0 |
0.7 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.6 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 |
0.4 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.1 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.0 |
0.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.3 |
GO:0002097 |
tRNA wobble base modification(GO:0002097) |
| 0.0 |
1.0 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.6 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
1.4 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.4 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of ubiquitin protein ligase activity(GO:1904667) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.1 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 |
0.7 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.6 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.2 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.2 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
0.5 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.3 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.1 |
GO:0097066 |
response to thyroid hormone(GO:0097066) |
| 0.0 |
0.4 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.9 |
GO:0032006 |
regulation of TOR signaling(GO:0032006) |
| 0.0 |
0.3 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.0 |
0.5 |
GO:0031960 |
response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 |
0.1 |
GO:0009446 |
putrescine metabolic process(GO:0009445) putrescine biosynthetic process(GO:0009446) |
| 0.0 |
0.7 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.1 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 |
0.1 |
GO:0090370 |
regulation of cholesterol efflux(GO:0010874) negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.2 |
GO:0034205 |
beta-amyloid formation(GO:0034205) regulation of beta-amyloid formation(GO:1902003) |
| 0.0 |
0.3 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.7 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.0 |
0.9 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.1 |
GO:0031953 |
negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 |
0.7 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.1 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.1 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
0.1 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.5 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 |
0.3 |
GO:0042026 |
protein refolding(GO:0042026) |
| 0.0 |
0.6 |
GO:0045727 |
positive regulation of translation(GO:0045727) |
| 0.0 |
0.4 |
GO:0014068 |
positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 |
0.0 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.0 |
0.1 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 |
0.1 |
GO:0043403 |
skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 |
0.1 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.0 |
0.5 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.3 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.2 |
GO:0035082 |
axoneme assembly(GO:0035082) |
| 0.0 |
0.4 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.7 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |