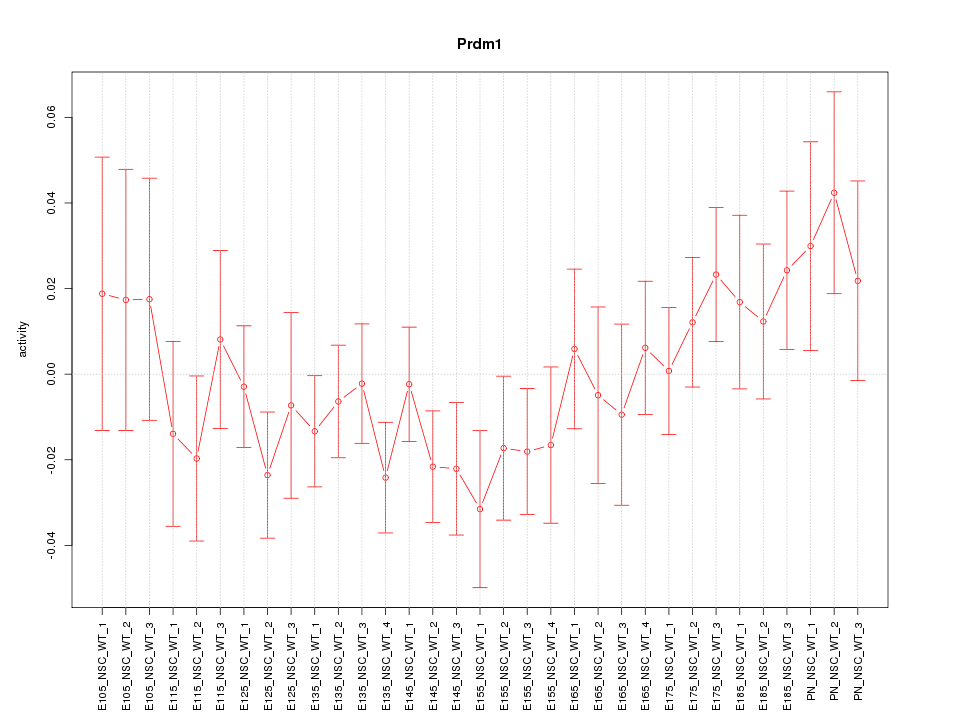
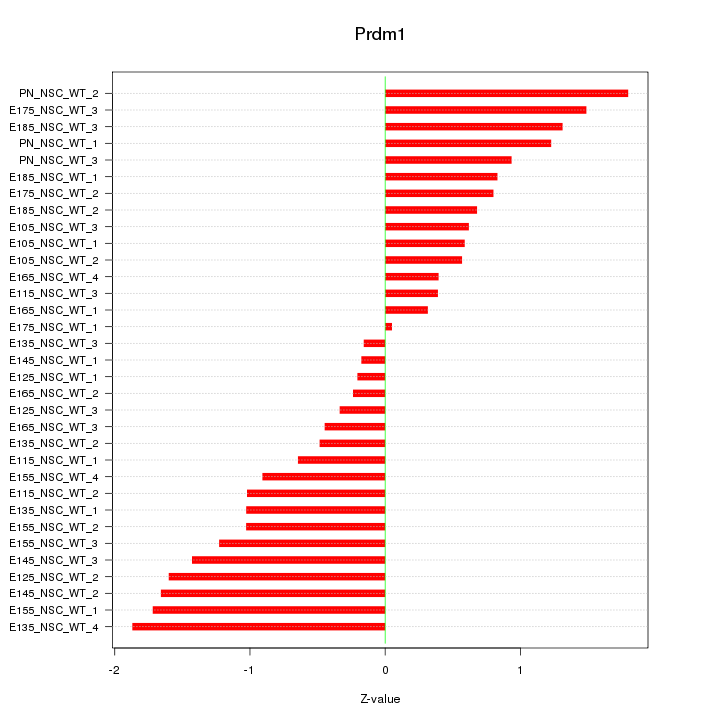
Motif ID: Prdm1
Z-value: 1.003

Transcription factors associated with Prdm1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Prdm1 | ENSMUSG00000038151.6 | Prdm1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 1.3 | 4.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 1.1 | 4.6 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 1.1 | 3.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.0 | 2.9 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.8 | 2.5 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.7 | 2.8 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.7 | 2.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.6 | 3.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.6 | 1.8 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.6 | 1.7 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.5 | 1.6 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.5 | 5.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.5 | 2.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.5 | 1.4 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.4 | 1.8 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.3 | 1.7 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.3 | 1.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 0.9 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.3 | 0.9 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) serotonin secretion by platelet(GO:0002554) |
| 0.3 | 0.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 2.3 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.2 | 0.7 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.2 | 1.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 2.6 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 0.9 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.2 | 3.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 2.0 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.2 | 1.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 0.8 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.2 | 3.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 1.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.2 | 0.7 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 0.8 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 0.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 1.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.8 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.4 | GO:0090335 | regulation of brown fat cell differentiation(GO:0090335) |
| 0.1 | 0.8 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 0.9 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 1.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 3.9 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.7 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 | 0.8 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.1 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.2 | GO:1990481 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 2.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.6 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 1.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 0.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.4 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 1.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 2.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.7 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.8 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.1 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 3.0 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.3 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.0 | 2.4 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.5 | 4.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.3 | 1.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.3 | 0.9 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 3.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 4.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 2.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 15.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 3.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.2 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 13.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 1.0 | 2.9 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.9 | 4.6 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.7 | 3.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.6 | 2.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.6 | 1.7 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.5 | 3.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.4 | 3.1 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.4 | 1.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 2.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 1.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 0.9 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 2.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 0.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 0.9 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.3 | 0.8 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.3 | 0.8 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 0.7 | GO:0032450 | oligo-1,6-glucosidase activity(GO:0004574) maltose alpha-glucosidase activity(GO:0032450) |
| 0.2 | 0.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 6.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 4.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 0.8 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 0.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.7 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 2.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.8 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) steroid hormone binding(GO:1990239) |
| 0.1 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 1.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 4.3 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.1 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 3.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 2.0 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 2.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.5 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.1 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 2.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0008198 | ATPase activator activity(GO:0001671) ferrous iron binding(GO:0008198) |
| 0.0 | 1.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |