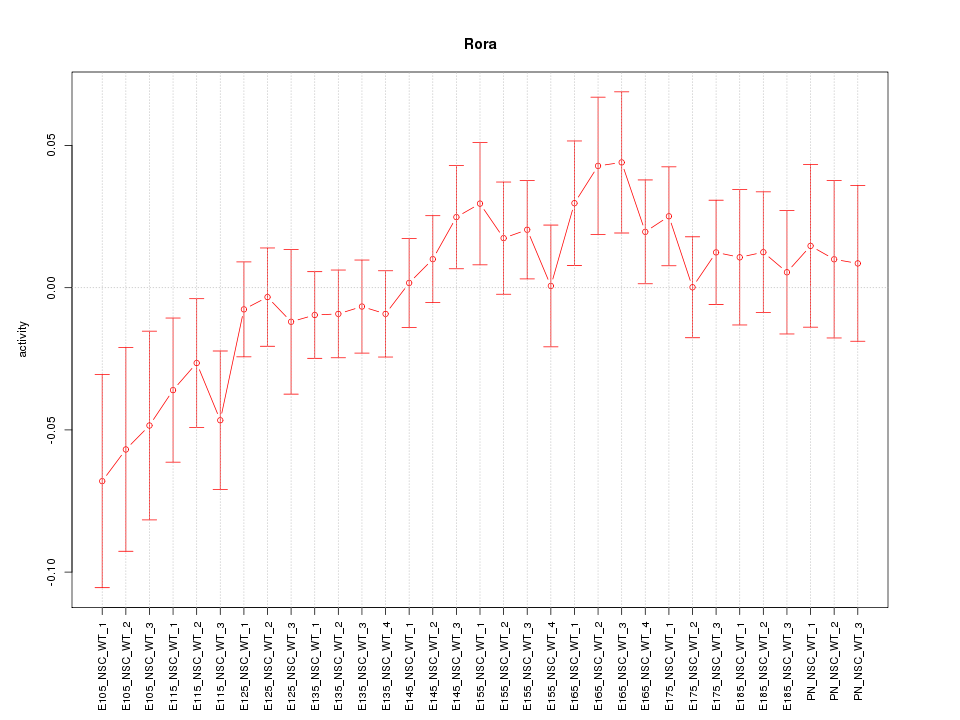
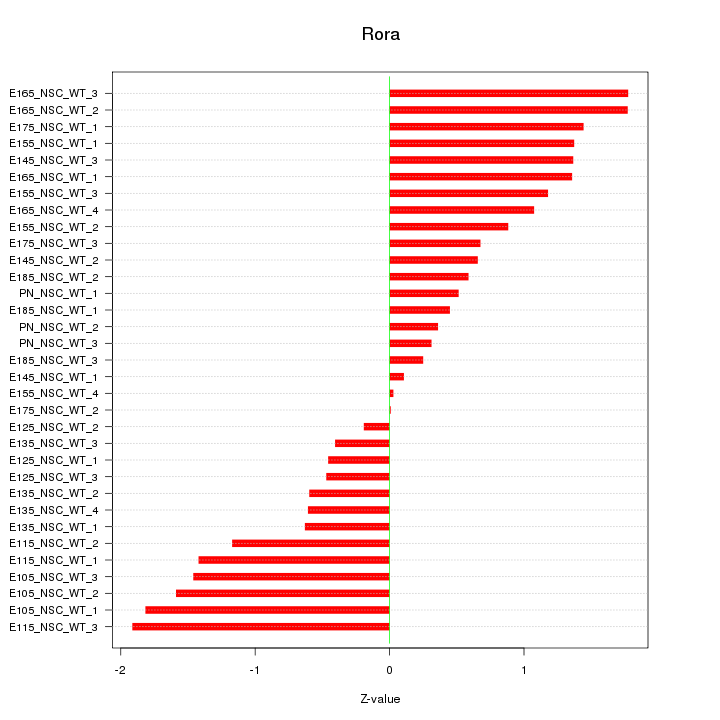
Motif ID: Rora
Z-value: 1.045
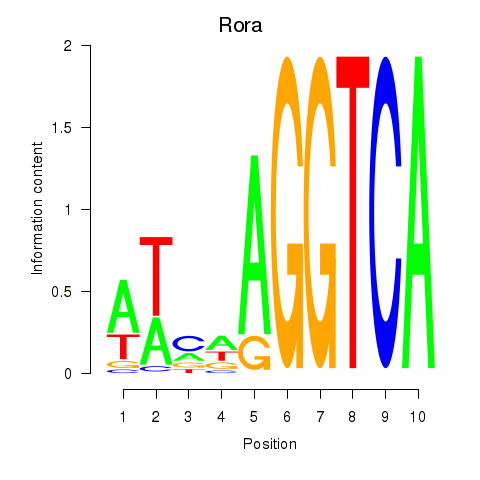
Transcription factors associated with Rora:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rora | ENSMUSG00000032238.11 | Rora |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rora | mm10_v2_chr9_+_68653761_68653786 | 0.37 | 3.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 1.7 | 5.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 1.4 | 4.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 1.4 | 4.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.2 | 5.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.0 | 3.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.0 | 3.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.9 | 4.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.8 | 2.4 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.8 | 3.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.7 | 2.2 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) |
| 0.7 | 6.6 | GO:0030432 | peristalsis(GO:0030432) |
| 0.7 | 2.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.6 | 2.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.6 | 1.9 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.6 | 5.5 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.6 | 4.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.6 | 3.6 | GO:1901524 | regulation of autophagosome maturation(GO:1901096) regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.6 | 3.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 2.7 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.5 | 3.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.5 | 2.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.5 | 3.6 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 | 3.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.4 | 3.5 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.4 | 2.4 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.4 | 1.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.4 | 3.0 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.4 | 2.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.4 | 2.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 2.6 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 2.1 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.3 | 1.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 2.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 2.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 1.4 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.2 | 1.4 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.2 | 0.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 0.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 4.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.1 | 0.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 12.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.5 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 2.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.3 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 2.6 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 | 2.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 3.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 2.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.3 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of protein glycosylation(GO:0060051) |
| 0.1 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 2.3 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.0 | 0.7 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 2.0 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.8 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 1.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 2.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.5 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.6 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 5.3 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 4.0 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.2 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 1.8 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.0 | 1.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 1.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 3.3 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 3.2 | GO:0015711 | organic anion transport(GO:0015711) |
| 0.0 | 2.0 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.0 | 2.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.9 | 6.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.7 | 3.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.7 | 4.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.6 | 2.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.4 | 2.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 1.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 1.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 5.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 2.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 3.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 1.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.7 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 2.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 3.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 8.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 4.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 7.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 7.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 7.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 2.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.8 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 11.1 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 11.9 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 6.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 9.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.2 | GO:0019841 | retinol binding(GO:0019841) |
| 2.2 | 6.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.0 | 3.0 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.8 | 2.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.6 | 3.8 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.6 | 3.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.6 | 6.1 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.6 | 6.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.6 | 2.4 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.6 | 3.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 3.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 2.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.5 | 4.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 3.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 7.4 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.4 | 5.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 2.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 2.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 2.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 3.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 4.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 2.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 2.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 4.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 4.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 2.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 2.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 0.9 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 1.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 2.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 3.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 4.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 4.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 3.0 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 6.8 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 2.5 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 2.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 4.6 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 3.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 3.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.6 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |