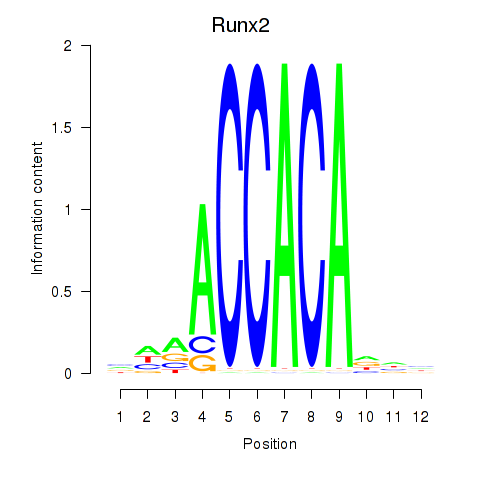
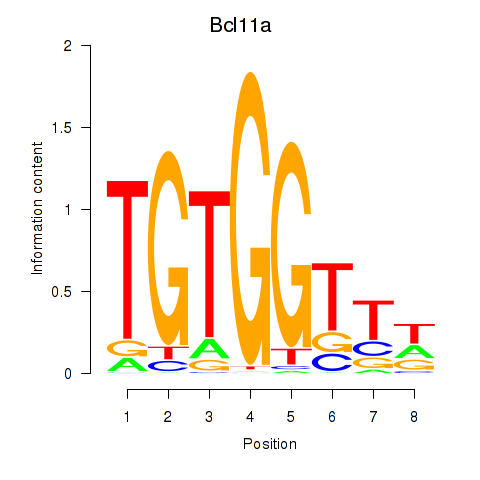
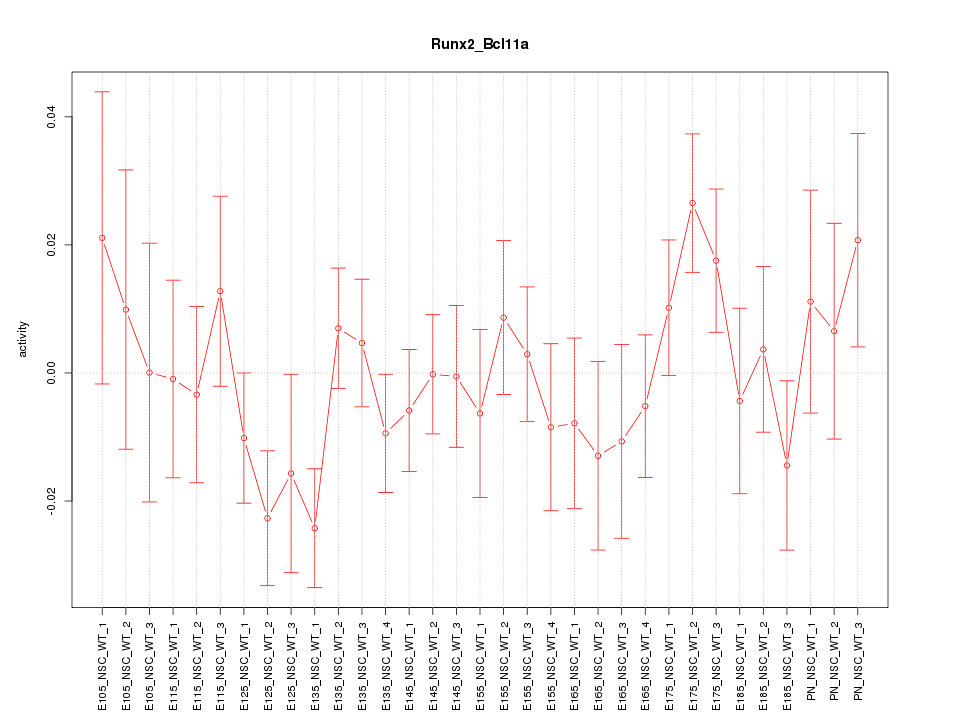
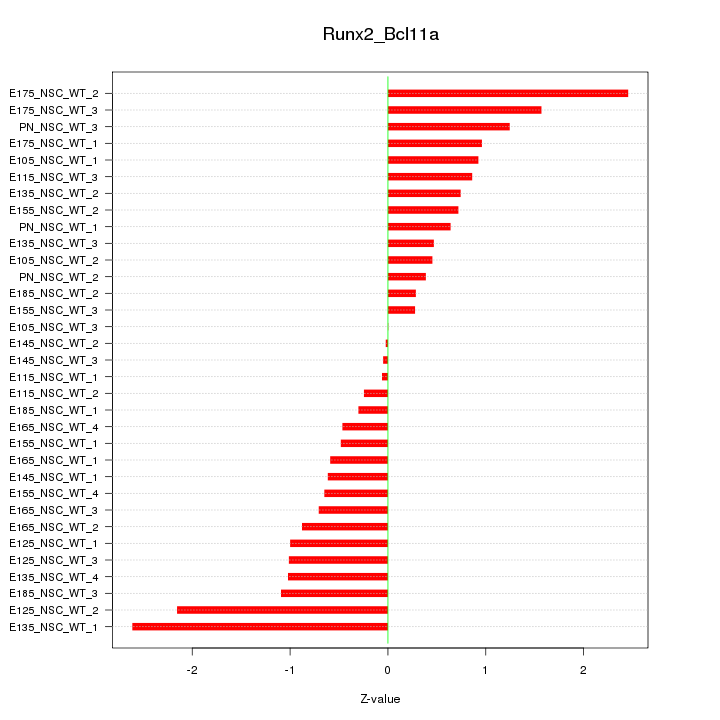
| 1.3 |
3.8 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 1.0 |
2.9 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.7 |
2.6 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.6 |
1.8 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.6 |
2.3 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.5 |
1.5 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.4 |
1.3 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.4 |
2.1 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.4 |
1.2 |
GO:0036292 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) DNA rewinding(GO:0036292) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 |
0.8 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.4 |
1.2 |
GO:0002034 |
angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.3 |
3.5 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.3 |
1.0 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.3 |
1.0 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.3 |
1.7 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 |
3.6 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.3 |
0.9 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.3 |
0.3 |
GO:0070858 |
negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.3 |
2.0 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.3 |
0.8 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 |
2.2 |
GO:1901387 |
positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.3 |
3.1 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.3 |
2.2 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 |
0.5 |
GO:0060769 |
positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.3 |
0.8 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 |
0.7 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.2 |
1.0 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.2 |
1.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.2 |
1.6 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.2 |
1.4 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.2 |
0.7 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.2 |
1.1 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) negative regulation of cytolysis(GO:0045918) |
| 0.2 |
1.9 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.2 |
0.6 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.2 |
0.8 |
GO:1900145 |
regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 |
1.2 |
GO:0021593 |
rhombomere morphogenesis(GO:0021593) |
| 0.2 |
1.6 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.2 |
1.0 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 |
0.6 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 |
0.6 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.2 |
2.0 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.2 |
1.0 |
GO:1902514 |
positive regulation of Rac protein signal transduction(GO:0035022) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 |
0.8 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 |
1.3 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 |
1.3 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 |
0.2 |
GO:1903463 |
regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 |
0.5 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) |
| 0.2 |
0.5 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.2 |
0.2 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.2 |
0.9 |
GO:0001705 |
ectoderm formation(GO:0001705) |
| 0.2 |
0.7 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.2 |
1.0 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
1.0 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 |
0.7 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 |
0.5 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.2 |
0.8 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.2 |
1.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 |
0.6 |
GO:0002339 |
B cell selection(GO:0002339) |
| 0.2 |
0.6 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.2 |
0.6 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 |
0.5 |
GO:0050929 |
cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 |
0.9 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 |
0.7 |
GO:0014054 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.1 |
1.1 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 |
0.6 |
GO:0010813 |
neuropeptide catabolic process(GO:0010813) |
| 0.1 |
1.0 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.4 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 |
0.5 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.3 |
GO:0070100 |
negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 |
0.4 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.1 |
0.9 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.5 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.1 |
0.5 |
GO:0010332 |
response to gamma radiation(GO:0010332) |
| 0.1 |
0.4 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
0.4 |
GO:0090035 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 |
1.2 |
GO:0071670 |
smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 |
0.4 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.6 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.4 |
GO:0042939 |
glutathione transport(GO:0034635) oligopeptide transmembrane transport(GO:0035672) xenobiotic transport(GO:0042908) tripeptide transport(GO:0042939) |
| 0.1 |
0.3 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 |
0.2 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
1.0 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.7 |
GO:0042756 |
drinking behavior(GO:0042756) |
| 0.1 |
0.8 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 |
0.6 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.3 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 |
0.4 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.8 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
0.7 |
GO:0060055 |
angiogenesis involved in wound healing(GO:0060055) |
| 0.1 |
0.3 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.1 |
2.4 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 |
0.4 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.1 |
0.7 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.1 |
1.3 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.1 |
3.4 |
GO:0070098 |
chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 |
0.4 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.1 |
0.9 |
GO:0033690 |
positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 |
0.6 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.9 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
1.0 |
GO:0021940 |
positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 |
0.6 |
GO:1904667 |
negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 |
2.3 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) |
| 0.1 |
0.2 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.1 |
0.4 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 |
0.5 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.4 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 |
0.3 |
GO:0006578 |
amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 |
0.4 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 |
0.8 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 |
0.2 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.1 |
0.5 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 |
1.7 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.3 |
GO:2001015 |
negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 |
0.5 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.1 |
0.2 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 |
0.6 |
GO:0035360 |
positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 |
0.4 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 |
0.4 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
1.0 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 |
0.7 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.4 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 |
0.8 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.1 |
1.5 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.3 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 |
0.9 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.1 |
0.7 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.3 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 |
0.2 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 |
1.6 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 |
0.2 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.3 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.1 |
0.5 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.3 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.1 |
3.7 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 |
1.0 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.1 |
0.4 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
0.3 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.1 |
0.5 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.1 |
0.2 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.1 |
1.2 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 |
0.4 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.1 |
0.2 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
0.3 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.2 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 |
0.3 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.1 |
1.0 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.1 |
0.2 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.1 |
1.0 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
0.5 |
GO:1901841 |
regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.1 |
0.4 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 |
0.2 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.1 |
2.0 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
1.2 |
GO:0022038 |
axonal fasciculation(GO:0007413) corpus callosum development(GO:0022038) |
| 0.1 |
0.3 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 |
0.6 |
GO:0033280 |
response to vitamin D(GO:0033280) |
| 0.1 |
0.3 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.2 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 |
0.2 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 |
0.2 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 |
0.3 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 |
0.2 |
GO:0060161 |
positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 |
0.2 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 |
0.1 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 |
0.3 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.1 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.0 |
0.2 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.4 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.0 |
0.2 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.2 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 |
0.2 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.2 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.1 |
GO:1901856 |
negative regulation of cellular respiration(GO:1901856) |
| 0.0 |
0.4 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.2 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 |
0.2 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.9 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
1.8 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.1 |
GO:0070346 |
positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 |
0.2 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 |
0.1 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 |
0.1 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 |
0.7 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 |
0.3 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
1.3 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.4 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.1 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 |
0.1 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 |
0.3 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.3 |
GO:0006693 |
prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 |
0.3 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0072362 |
regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 |
1.2 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 |
0.1 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 |
0.2 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.3 |
GO:0097084 |
vascular smooth muscle cell development(GO:0097084) |
| 0.0 |
0.1 |
GO:0015819 |
lysine transport(GO:0015819) |
| 0.0 |
0.5 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.0 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.2 |
GO:1900048 |
positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.0 |
0.1 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 |
0.1 |
GO:0042940 |
D-amino acid transport(GO:0042940) |
| 0.0 |
1.0 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.3 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.0 |
0.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.4 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.1 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 |
0.3 |
GO:0045776 |
negative regulation of blood pressure(GO:0045776) |
| 0.0 |
0.1 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 |
0.1 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 |
0.6 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.3 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.4 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
2.8 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.3 |
GO:0010574 |
regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.0 |
0.1 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.5 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0001780 |
neutrophil homeostasis(GO:0001780) |
| 0.0 |
0.5 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.2 |
GO:0070571 |
negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 |
0.5 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.0 |
0.5 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.4 |
GO:0001779 |
natural killer cell differentiation(GO:0001779) |
| 0.0 |
0.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.4 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.0 |
0.4 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.2 |
GO:0050667 |
homocysteine metabolic process(GO:0050667) |
| 0.0 |
0.3 |
GO:0006893 |
Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 |
0.2 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.6 |
GO:0033137 |
negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 |
0.1 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.0 |
0.5 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.1 |
GO:0007548 |
sex differentiation(GO:0007548) |
| 0.0 |
0.6 |
GO:0019915 |
lipid storage(GO:0019915) |
| 0.0 |
0.1 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.1 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.0 |
0.1 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 |
0.2 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.1 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.8 |
GO:0050810 |
regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 |
0.7 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.1 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.1 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.1 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.0 |
0.1 |
GO:0042148 |
strand invasion(GO:0042148) |
| 0.0 |
0.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.1 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 |
0.4 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.1 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 |
0.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.5 |
GO:0035082 |
axoneme assembly(GO:0035082) |
| 0.0 |
0.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.6 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.4 |
GO:0007094 |
mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 |
0.0 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
0.1 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.7 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.2 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 |
0.1 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.0 |
0.2 |
GO:0009263 |
deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 |
0.0 |
GO:0045617 |
negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 |
0.2 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.2 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.7 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.2 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
0.1 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.1 |
GO:1903894 |
regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 |
0.1 |
GO:0034351 |
negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 |
0.0 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.4 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |