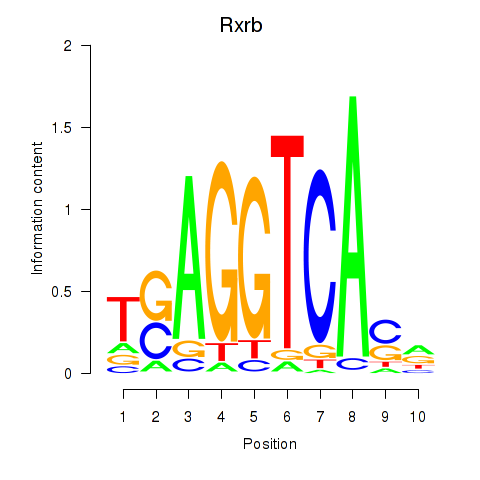
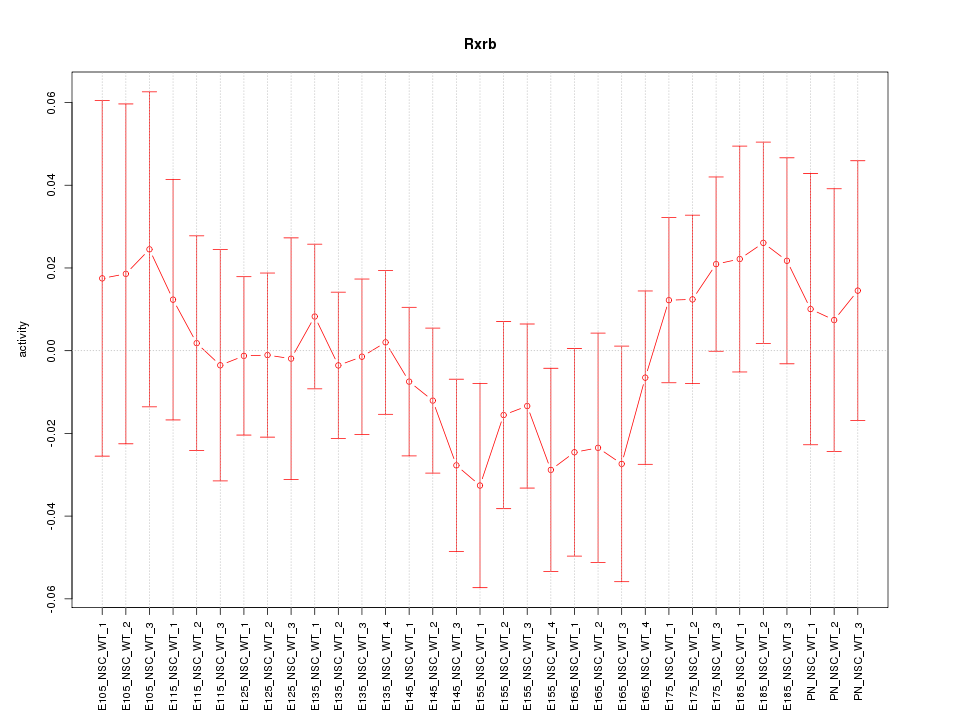
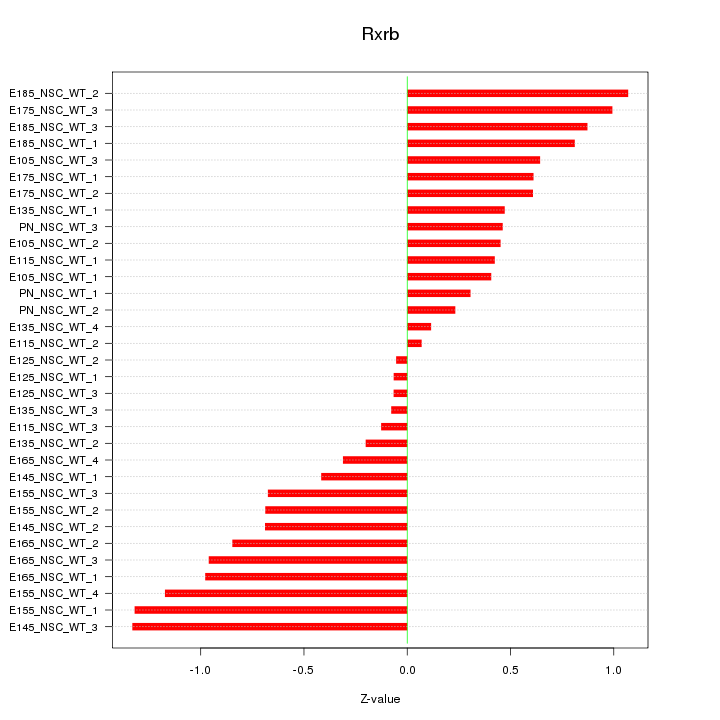
| 0.6 |
1.9 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.6 |
1.7 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.4 |
2.6 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 |
1.9 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.4 |
1.8 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 |
1.7 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.3 |
1.0 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 |
1.3 |
GO:1903463 |
mitotic cell cycle phase(GO:0098763) regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 |
1.2 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 |
1.2 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 |
0.5 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 |
0.2 |
GO:0055118 |
negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.2 |
1.0 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
4.7 |
GO:0071398 |
cellular response to fatty acid(GO:0071398) |
| 0.2 |
2.4 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.2 |
0.6 |
GO:0097494 |
regulation of vesicle size(GO:0097494) |
| 0.2 |
1.1 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.4 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 |
1.1 |
GO:0061084 |
negative regulation of protein refolding(GO:0061084) |
| 0.1 |
1.1 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 0.1 |
0.4 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.1 |
0.6 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 |
0.8 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.1 |
1.9 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
0.6 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
0.4 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) |
| 0.1 |
1.8 |
GO:0051894 |
integrin activation(GO:0033622) positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 |
0.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.6 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.1 |
1.7 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 |
0.8 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
0.4 |
GO:1903753 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) hard palate development(GO:0060022) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 |
0.7 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 |
0.3 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.4 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.2 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 |
1.4 |
GO:0006760 |
folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 |
0.1 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 |
0.2 |
GO:0034115 |
negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
1.6 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.8 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.0 |
0.7 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.5 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.6 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
1.5 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 |
1.9 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.0 |
0.5 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.6 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
0.7 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.3 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.1 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
1.5 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.2 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
1.0 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |