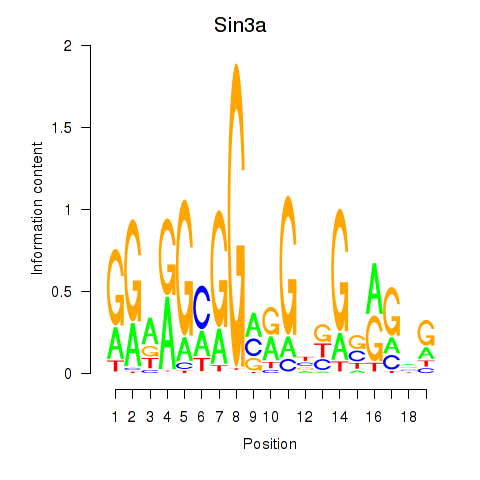
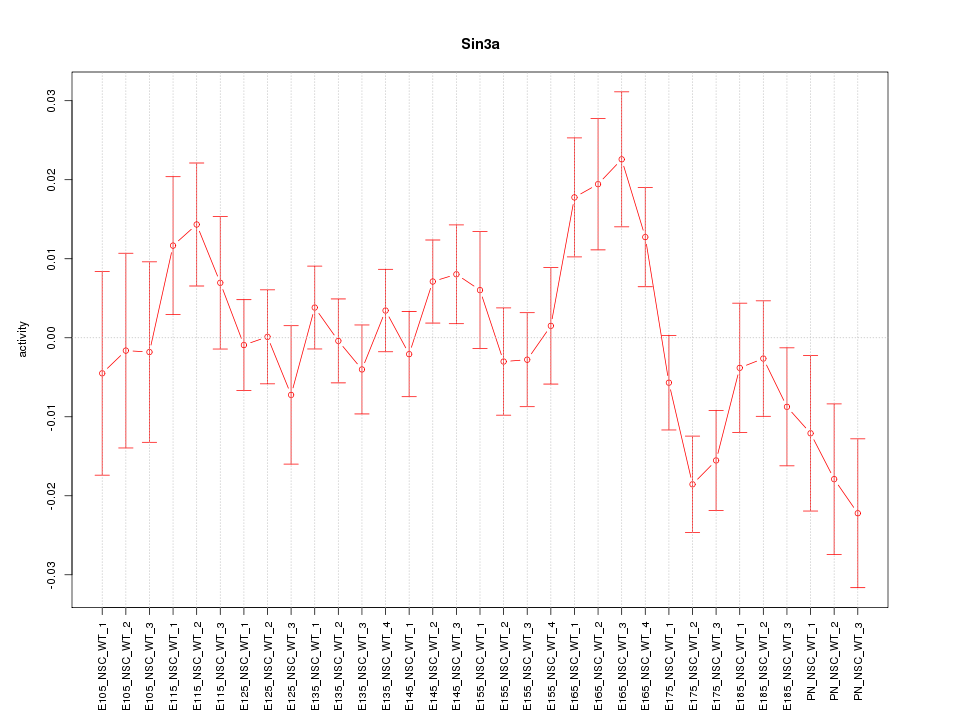
| 1.7 |
5.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 1.3 |
4.0 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.8 |
4.2 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.8 |
2.3 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.7 |
7.1 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.7 |
2.0 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.7 |
10.1 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 |
3.9 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.6 |
2.4 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.5 |
8.7 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.5 |
0.5 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.5 |
1.5 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.5 |
1.5 |
GO:0031990 |
mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.5 |
3.0 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.5 |
1.4 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.5 |
4.6 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.5 |
0.9 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.5 |
1.8 |
GO:0071692 |
protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.5 |
1.4 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.4 |
0.9 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.4 |
1.8 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.4 |
1.7 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.4 |
2.6 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 |
0.8 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.4 |
1.6 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.4 |
1.2 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.4 |
1.2 |
GO:0031622 |
positive regulation of fever generation(GO:0031622) |
| 0.4 |
2.0 |
GO:0021699 |
cerebellar cortex maturation(GO:0021699) |
| 0.4 |
1.2 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.4 |
0.8 |
GO:1902661 |
regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.4 |
1.2 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.4 |
1.1 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.4 |
3.3 |
GO:0061038 |
uterus morphogenesis(GO:0061038) |
| 0.4 |
0.7 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.4 |
1.1 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 |
0.7 |
GO:0002329 |
pre-B cell differentiation(GO:0002329) |
| 0.4 |
1.4 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.3 |
1.4 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.3 |
1.4 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.3 |
1.3 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.3 |
1.0 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 |
1.3 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 |
1.0 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 |
0.9 |
GO:0010751 |
negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.3 |
2.8 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.3 |
1.2 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 |
0.6 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 |
0.9 |
GO:1901536 |
regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.3 |
0.9 |
GO:0061303 |
cornea development in camera-type eye(GO:0061303) |
| 0.3 |
3.3 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) cellular response to magnesium ion(GO:0071286) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 |
0.6 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 |
2.3 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.3 |
1.4 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 |
0.9 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.3 |
0.6 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) |
| 0.3 |
0.5 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.3 |
2.7 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.3 |
0.3 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.3 |
0.3 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.3 |
0.8 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.3 |
0.3 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 |
2.3 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.3 |
0.8 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 |
1.0 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.3 |
1.0 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.3 |
1.3 |
GO:1900109 |
regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.3 |
1.0 |
GO:0071415 |
cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.3 |
3.8 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.3 |
0.8 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 |
2.5 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 |
3.2 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
0.7 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.2 |
0.7 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.2 |
1.5 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.2 |
1.9 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 |
3.1 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.2 |
1.4 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 |
0.5 |
GO:0048048 |
embryonic eye morphogenesis(GO:0048048) |
| 0.2 |
1.9 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.2 |
0.9 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.2 |
1.2 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.2 |
0.9 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.2 |
0.9 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.2 |
2.7 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.2 |
0.9 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) histidine transport(GO:0015817) |
| 0.2 |
0.9 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.2 |
0.7 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.2 |
0.7 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.2 |
0.7 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.2 |
0.2 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 |
0.4 |
GO:2001015 |
negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.2 |
0.7 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.2 |
0.7 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 |
0.7 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 |
0.7 |
GO:0016332 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 |
1.9 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 |
0.9 |
GO:0046958 |
nonassociative learning(GO:0046958) |
| 0.2 |
4.1 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.2 |
1.5 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.2 |
1.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.2 |
2.3 |
GO:0090394 |
negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.2 |
0.8 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.2 |
0.8 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 |
0.6 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.2 |
0.6 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.2 |
0.4 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.2 |
0.8 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.2 |
1.6 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.2 |
0.8 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.2 |
1.6 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.2 |
0.8 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.2 |
0.8 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 |
2.1 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.2 |
0.4 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.2 |
2.6 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.2 |
1.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 |
1.3 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.2 |
2.0 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 |
0.5 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.2 |
1.5 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 |
0.7 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.2 |
0.5 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.2 |
0.4 |
GO:0071880 |
adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.2 |
6.2 |
GO:0021680 |
cerebellar Purkinje cell layer development(GO:0021680) |
| 0.2 |
0.4 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.2 |
1.2 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 |
1.7 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 |
0.3 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 |
0.3 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 |
1.6 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.2 |
1.0 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.2 |
2.0 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 |
0.7 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 |
0.7 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 |
1.5 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 |
0.8 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 |
2.0 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 |
0.5 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.2 |
1.0 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.2 |
0.3 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.2 |
1.3 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.2 |
0.7 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 |
0.8 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.2 |
1.3 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 |
0.2 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.2 |
0.8 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.2 |
1.5 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 |
1.0 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.2 |
0.6 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.2 |
0.8 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 |
1.3 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 |
1.1 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.2 |
0.6 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 |
0.5 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.2 |
0.2 |
GO:0051570 |
regulation of histone H3-K9 methylation(GO:0051570) |
| 0.2 |
0.5 |
GO:0003211 |
cardiac ventricle formation(GO:0003211) |
| 0.2 |
0.5 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 |
0.5 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 |
0.8 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.2 |
1.2 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.2 |
1.4 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.2 |
0.5 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.1 |
0.6 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.4 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 |
0.6 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 |
0.3 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 |
5.1 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.1 |
0.4 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
0.9 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.1 |
0.4 |
GO:0034241 |
positive regulation of macrophage fusion(GO:0034241) regulation of osteoclast proliferation(GO:0090289) |
| 0.1 |
0.1 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 |
2.1 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 |
1.1 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.1 |
GO:0098910 |
regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.1 |
0.1 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.1 |
3.4 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 |
0.5 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
1.8 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.1 |
0.4 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.1 |
0.4 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.7 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
1.2 |
GO:2000394 |
positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 |
0.4 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.1 |
1.3 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 |
0.7 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.4 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
0.5 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 |
0.6 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.5 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.9 |
GO:0010825 |
positive regulation of centrosome duplication(GO:0010825) |
| 0.1 |
0.6 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
1.8 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.5 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.6 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.1 |
1.0 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
0.4 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
2.2 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
2.0 |
GO:0086018 |
SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.1 |
0.4 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 |
0.7 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
0.1 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 |
0.7 |
GO:0045072 |
regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 |
1.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.1 |
0.6 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.1 |
GO:2001268 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 |
0.6 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 |
0.4 |
GO:0097151 |
terminal button organization(GO:0072553) positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 |
0.4 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 |
0.6 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 |
1.3 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
0.2 |
GO:0043950 |
positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 |
1.1 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 |
0.1 |
GO:2000642 |
negative regulation of vacuolar transport(GO:1903336) regulation of early endosome to late endosome transport(GO:2000641) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.5 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
0.2 |
GO:2000664 |
positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 |
0.8 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.1 |
0.5 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.1 |
0.3 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.4 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 |
0.2 |
GO:0034969 |
histone arginine methylation(GO:0034969) |
| 0.1 |
0.6 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.3 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 |
0.5 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
2.7 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 |
0.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.6 |
GO:1901339 |
regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 |
0.3 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.1 |
0.8 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
1.3 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.1 |
0.7 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.1 |
0.1 |
GO:0019732 |
antifungal humoral response(GO:0019732) |
| 0.1 |
1.6 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
0.6 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 |
0.7 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.4 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
0.4 |
GO:0034392 |
negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 |
0.5 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
1.0 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.4 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.1 |
0.8 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.1 |
0.4 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
2.4 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
0.5 |
GO:0021592 |
fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.1 |
0.5 |
GO:0045918 |
negative regulation of cytolysis(GO:0045918) |
| 0.1 |
0.6 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.1 |
0.8 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.3 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.3 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.9 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 |
4.7 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.1 |
1.4 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.1 |
0.2 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.8 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 |
0.6 |
GO:0090306 |
spindle assembly involved in meiosis(GO:0090306) |
| 0.1 |
0.3 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.1 |
0.2 |
GO:0046098 |
regulation of primitive erythrocyte differentiation(GO:0010725) guanine metabolic process(GO:0046098) |
| 0.1 |
3.2 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
0.2 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.1 |
1.1 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.1 |
1.8 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.1 |
0.6 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.7 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 |
2.5 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.1 |
0.2 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 |
0.9 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.4 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.2 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.1 |
0.2 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 |
0.6 |
GO:0071361 |
detection of calcium ion(GO:0005513) cellular response to ethanol(GO:0071361) |
| 0.1 |
0.2 |
GO:0072675 |
multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 |
0.6 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 |
0.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.3 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 |
0.5 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.1 |
0.5 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.3 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.1 |
GO:0071879 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 |
0.3 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.1 |
0.2 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.1 |
0.2 |
GO:0060743 |
prostate gland stromal morphogenesis(GO:0060741) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
0.1 |
GO:0060994 |
regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 |
0.4 |
GO:0070100 |
negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 |
0.4 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.8 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
0.6 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.1 |
0.4 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.3 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.1 |
0.2 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.1 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
1.3 |
GO:0021895 |
cerebral cortex neuron differentiation(GO:0021895) |
| 0.1 |
0.3 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 |
1.2 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.1 |
GO:0001787 |
natural killer cell proliferation(GO:0001787) |
| 0.1 |
0.1 |
GO:0070560 |
protein secretion by platelet(GO:0070560) |
| 0.1 |
0.7 |
GO:0044331 |
cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 |
0.4 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.3 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.1 |
0.4 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.1 |
0.4 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 |
0.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.3 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 |
0.1 |
GO:0021559 |
trigeminal nerve development(GO:0021559) |
| 0.1 |
0.4 |
GO:0044387 |
negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 |
0.5 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.5 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.3 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
2.1 |
GO:1902668 |
negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 |
0.7 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.6 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 |
0.5 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.1 |
0.2 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 |
0.3 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.1 |
GO:0000966 |
RNA 5'-end processing(GO:0000966) |
| 0.1 |
0.3 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
0.9 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 |
0.4 |
GO:1903056 |
mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.1 |
GO:0048148 |
behavioral response to cocaine(GO:0048148) |
| 0.1 |
0.4 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.1 |
0.2 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.1 |
0.2 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.1 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.1 |
0.2 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 |
0.2 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.1 |
1.1 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.1 |
0.7 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.5 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.2 |
GO:0060368 |
regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 |
1.0 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.1 |
0.2 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.4 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 |
0.2 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) |
| 0.1 |
0.7 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 |
0.5 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.1 |
0.2 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.3 |
GO:2000288 |
positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 |
0.2 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.1 |
0.3 |
GO:0023019 |
signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.1 |
0.5 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
0.5 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.1 |
0.6 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 |
0.6 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 |
0.2 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
0.2 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 |
0.5 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
1.2 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
1.0 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
1.2 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.4 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.1 |
0.8 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.1 |
0.3 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.7 |
GO:0014068 |
positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 |
1.7 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.1 |
0.2 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.9 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.1 |
0.2 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 |
1.3 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 |
1.0 |
GO:0031061 |
negative regulation of histone methylation(GO:0031061) |
| 0.1 |
1.0 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
1.5 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.1 |
0.4 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 |
1.1 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.6 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.1 |
0.2 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.6 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
0.4 |
GO:0055090 |
acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 |
0.9 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 |
0.4 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.6 |
GO:0032418 |
lysosome localization(GO:0032418) |
| 0.0 |
0.1 |
GO:0097503 |
sialylation(GO:0097503) protein sialylation(GO:1990743) |
| 0.0 |
0.1 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.2 |
GO:0055009 |
extraocular skeletal muscle development(GO:0002074) atrial cardiac muscle tissue development(GO:0003228) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) atrial cardiac muscle tissue morphogenesis(GO:0055009) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) endodermal digestive tract morphogenesis(GO:0061031) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 |
0.5 |
GO:0072673 |
lamellipodium morphogenesis(GO:0072673) |
| 0.0 |
0.2 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 |
0.3 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
1.0 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.5 |
GO:0051000 |
positive regulation of nitric-oxide synthase activity(GO:0051000) |
| 0.0 |
0.4 |
GO:0042790 |
transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 |
0.2 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.0 |
GO:2000468 |
regulation of peroxidase activity(GO:2000468) |
| 0.0 |
0.7 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.2 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 |
0.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.4 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.4 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.5 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.1 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 |
6.4 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.1 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 |
0.4 |
GO:0001960 |
negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 |
0.4 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.1 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 |
0.2 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
1.3 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.6 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.3 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.3 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.2 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.1 |
GO:2001258 |
negative regulation of cation channel activity(GO:2001258) |
| 0.0 |
0.2 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 |
0.3 |
GO:0045921 |
positive regulation of exocytosis(GO:0045921) |
| 0.0 |
2.4 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
3.9 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
5.1 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.1 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.5 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.4 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.3 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
0.1 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.3 |
GO:0071028 |
nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.3 |
GO:1901621 |
regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
2.0 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.5 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
1.2 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.0 |
0.2 |
GO:0007603 |
phototransduction, visible light(GO:0007603) |
| 0.0 |
0.2 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.0 |
0.3 |
GO:0033127 |
regulation of histone phosphorylation(GO:0033127) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
1.3 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.4 |
GO:0019243 |
methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.1 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.0 |
0.1 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.0 |
0.6 |
GO:0071806 |
intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 |
0.8 |
GO:0048255 |
mRNA stabilization(GO:0048255) |
| 0.0 |
0.0 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 |
0.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.0 |
0.4 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.3 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.0 |
0.0 |
GO:0043921 |
modulation by host of viral transcription(GO:0043921) modulation of transcription in other organism involved in symbiotic interaction(GO:0052312) modulation by host of symbiont transcription(GO:0052472) |
| 0.0 |
0.3 |
GO:0060546 |
negative regulation of necroptotic process(GO:0060546) |
| 0.0 |
0.3 |
GO:0010390 |
histone monoubiquitination(GO:0010390) |
| 0.0 |
0.2 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.0 |
0.9 |
GO:0072384 |
organelle transport along microtubule(GO:0072384) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.3 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.4 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.3 |
GO:0090102 |
cochlea development(GO:0090102) |
| 0.0 |
0.3 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.5 |
GO:0035116 |
embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 |
0.1 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.1 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.0 |
0.4 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.0 |
0.3 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.1 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 |
0.3 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.4 |
GO:0090280 |
positive regulation of calcium ion import(GO:0090280) |
| 0.0 |
0.1 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.0 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 |
0.3 |
GO:1903541 |
regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) |
| 0.0 |
0.1 |
GO:1902036 |
regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 |
0.1 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 |
0.1 |
GO:1903867 |
chorion development(GO:0060717) extraembryonic membrane development(GO:1903867) |
| 0.0 |
0.2 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.1 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.1 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.5 |
GO:0034243 |
regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 |
0.0 |
GO:0048715 |
negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 |
0.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
0.3 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.0 |
0.2 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 |
0.7 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
1.5 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.3 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.1 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.3 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.2 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.7 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.1 |
GO:0090169 |
regulation of spindle assembly(GO:0090169) |
| 0.0 |
0.3 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.1 |
GO:0060317 |
cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 |
0.0 |
GO:0043970 |
histone H3-K9 acetylation(GO:0043970) |
| 0.0 |
0.4 |
GO:0048247 |
lymphocyte chemotaxis(GO:0048247) |
| 0.0 |
0.1 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.2 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.1 |
GO:0019249 |
lactate metabolic process(GO:0006089) lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 |
0.3 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.0 |
GO:0003057 |
regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 |
1.1 |
GO:1903749 |
positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 |
0.4 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.2 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.1 |
GO:0003419 |
growth plate cartilage chondrocyte proliferation(GO:0003419) regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.0 |
0.7 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
0.7 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.1 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.1 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.0 |
0.1 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.0 |
GO:0042249 |
establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 |
0.2 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.3 |
GO:0019884 |
antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 |
0.2 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.8 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.1 |
GO:0044090 |
positive regulation of vacuole organization(GO:0044090) |
| 0.0 |
0.1 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.1 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.0 |
0.2 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.2 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.1 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.1 |
GO:0045823 |
positive regulation of heart contraction(GO:0045823) |
| 0.0 |
0.2 |
GO:0016239 |
positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.0 |
GO:0030011 |
maintenance of cell polarity(GO:0030011) |
| 0.0 |
0.1 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.1 |
GO:0045932 |
negative regulation of muscle contraction(GO:0045932) |
| 0.0 |
0.3 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.0 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 |
0.1 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 |
0.2 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.1 |
GO:0044872 |
lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 |
0.1 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.2 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.0 |
GO:0034397 |
telomere localization(GO:0034397) |
| 0.0 |
0.0 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.2 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.0 |
GO:0032612 |
interleukin-1 production(GO:0032612) |
| 0.0 |
0.0 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.5 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.0 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.0 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 |
0.2 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.1 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.1 |
GO:2000352 |
negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 |
0.2 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.1 |
GO:0065005 |
protein-lipid complex assembly(GO:0065005) |