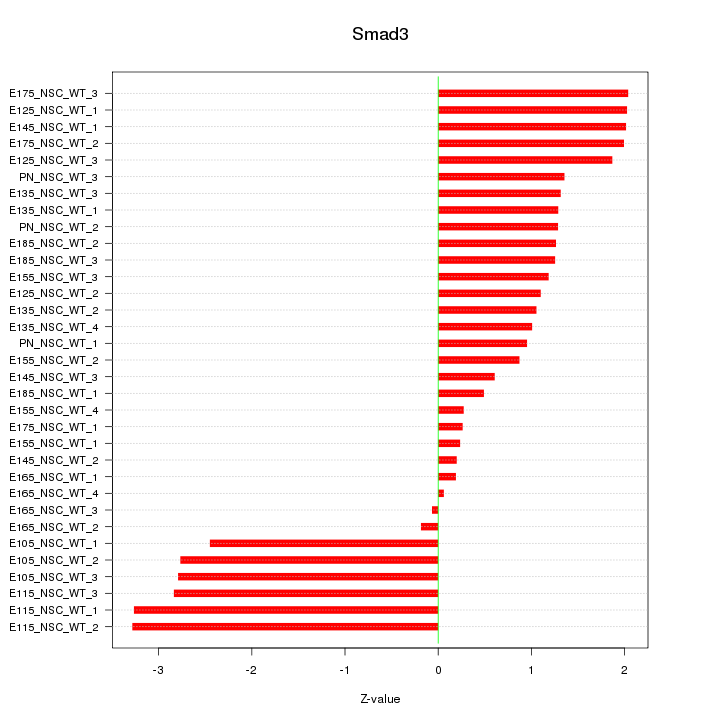
Motif ID: Smad3
Z-value: 1.634
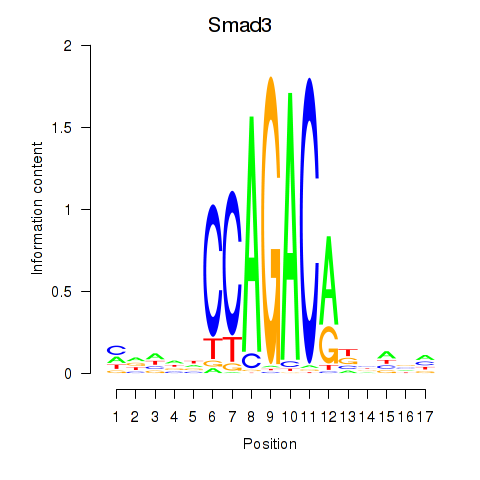
Transcription factors associated with Smad3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Smad3 | ENSMUSG00000032402.6 | Smad3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad3 | mm10_v2_chr9_-_63711969_63711994 | -0.72 | 2.5e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 2.5 | 7.6 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 2.2 | 6.6 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) |
| 2.1 | 6.4 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 1.9 | 5.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 1.5 | 4.5 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.4 | 26.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.2 | 5.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 1.0 | 3.1 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.9 | 3.6 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) peptidyl-aspartic acid modification(GO:0018197) |
| 0.8 | 90.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.7 | 8.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 3.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 2.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 11.0 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.3 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.7 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 2.9 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 2.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 2.5 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.3 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 7.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 2.6 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 0.8 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.1 | 2.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 2.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 1.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 3.7 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 15.6 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 3.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 11.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.1 | 82.6 | GO:0000786 | nucleosome(GO:0000786) |
| 1.0 | 3.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.3 | 4.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 6.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 3.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 3.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 5.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 39.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 7.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 14.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.0 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 5.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.5 | 4.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.2 | 11.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.1 | 6.4 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.9 | 6.6 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 2.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.4 | 3.6 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.3 | 8.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 3.5 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 1.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 0.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 3.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 6.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 1.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 24.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 5.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.6 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 3.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 46.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 3.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.8 | GO:0035925 | translation repressor activity, nucleic acid binding(GO:0000900) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 62.8 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 3.9 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |