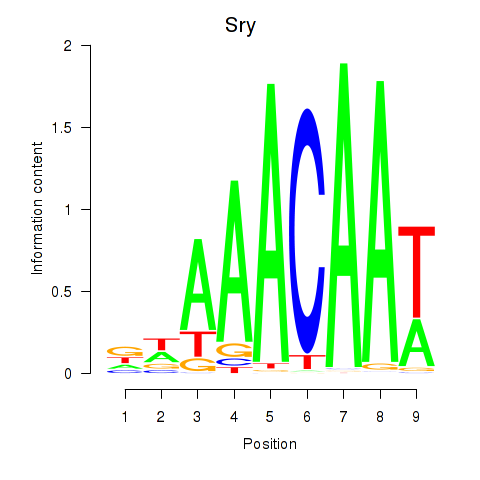
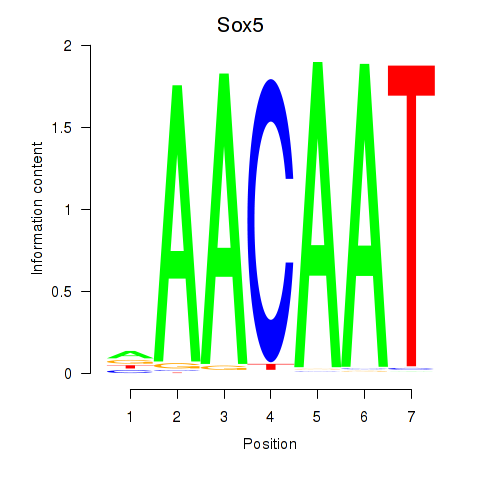
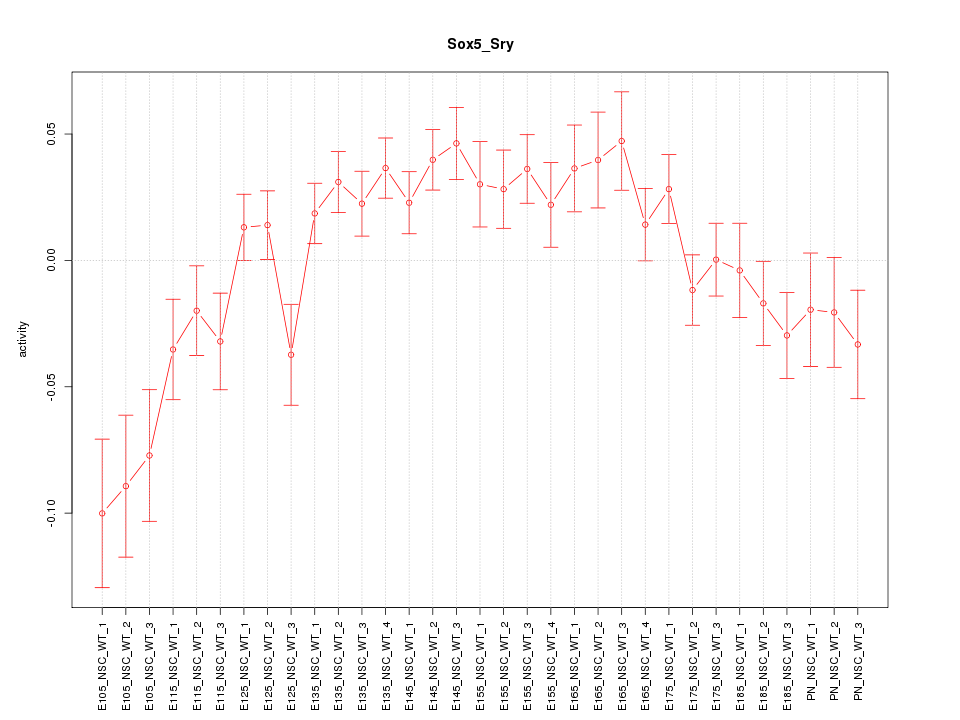
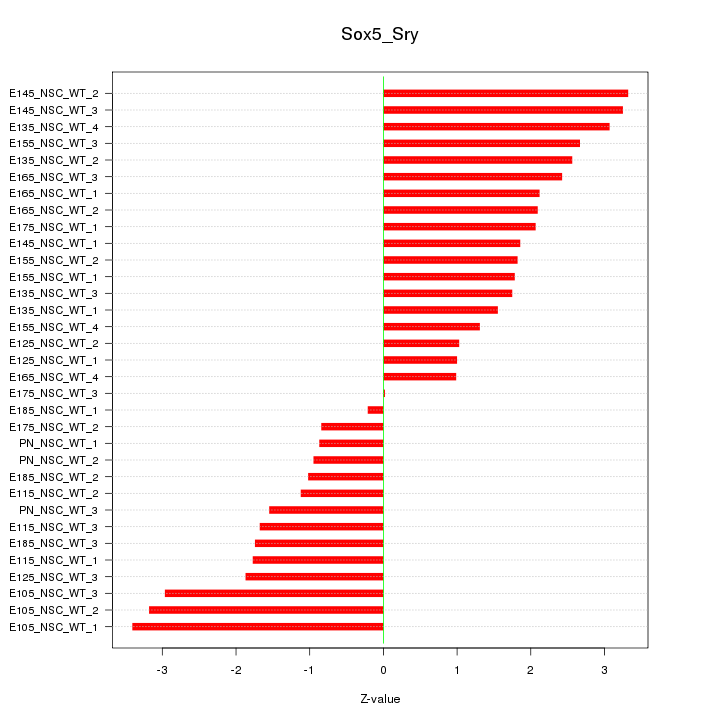
| 14.6 |
58.5 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 12.8 |
38.5 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 7.8 |
23.3 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 4.3 |
12.8 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.9 |
31.8 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.5 |
17.3 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 2.1 |
8.3 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 2.1 |
20.8 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 1.9 |
27.2 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.7 |
10.4 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 1.6 |
11.3 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.3 |
5.2 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.3 |
2.5 |
GO:0046543 |
development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 1.1 |
7.8 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 1.1 |
5.6 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 1.0 |
8.2 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 1.0 |
3.9 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.0 |
3.8 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.9 |
2.7 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.9 |
2.7 |
GO:1900039 |
positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.9 |
5.3 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.9 |
3.5 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.8 |
4.2 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.8 |
15.0 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.8 |
3.3 |
GO:0061428 |
embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.8 |
3.2 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 |
2.3 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.8 |
3.9 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.8 |
3.9 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.7 |
3.0 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.7 |
7.1 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 |
5.7 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.7 |
1.4 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.7 |
4.7 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.6 |
3.2 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.6 |
8.9 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.6 |
6.1 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.6 |
4.8 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.6 |
7.5 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.6 |
2.8 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.5 |
1.0 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 0.5 |
4.0 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.5 |
4.5 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 |
18.5 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.5 |
2.0 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.5 |
2.4 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 |
3.9 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.4 |
1.7 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.4 |
2.9 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.4 |
2.4 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 |
10.3 |
GO:0030204 |
chondroitin sulfate metabolic process(GO:0030204) |
| 0.4 |
4.2 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 |
5.6 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.4 |
2.9 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.4 |
4.7 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.3 |
3.1 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.3 |
1.6 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 |
3.1 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.3 |
8.6 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.3 |
6.0 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.3 |
11.9 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.3 |
5.4 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 |
1.4 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.3 |
1.4 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 |
3.5 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.3 |
2.6 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.3 |
0.5 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.2 |
2.2 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 |
1.2 |
GO:0008655 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.2 |
0.9 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.2 |
1.6 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 |
0.7 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.2 |
1.4 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.2 |
0.8 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 |
1.4 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 |
0.6 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 |
0.7 |
GO:0008078 |
mesodermal cell migration(GO:0008078) |
| 0.2 |
1.3 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 |
4.1 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 |
1.1 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.2 |
1.7 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.2 |
0.5 |
GO:0017085 |
response to insecticide(GO:0017085) |
| 0.2 |
1.1 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.2 |
0.9 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.4 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
1.5 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
1.8 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
0.4 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
2.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
3.6 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
1.0 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 |
1.2 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.1 |
3.9 |
GO:0030890 |
positive regulation of B cell proliferation(GO:0030890) |
| 0.1 |
1.3 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.6 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
0.9 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
4.9 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.1 |
0.6 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
1.9 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 |
1.4 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
7.9 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.1 |
0.6 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
1.0 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
0.9 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.1 |
1.8 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.1 |
6.3 |
GO:0021954 |
central nervous system neuron development(GO:0021954) |
| 0.1 |
0.8 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.1 |
6.2 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.8 |
GO:0051447 |
negative regulation of meiotic cell cycle(GO:0051447) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.6 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
2.4 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.1 |
2.5 |
GO:1900449 |
regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 |
3.7 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.1 |
3.4 |
GO:0051784 |
negative regulation of mitotic nuclear division(GO:0045839) negative regulation of nuclear division(GO:0051784) |
| 0.1 |
2.0 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.5 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.1 |
0.5 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 |
2.1 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
1.3 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
1.2 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
1.1 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.1 |
0.4 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
1.4 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
1.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
1.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.9 |
GO:0072348 |
sulfur compound transport(GO:0072348) |
| 0.0 |
0.3 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 |
1.5 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.8 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.7 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.2 |
GO:0089711 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.3 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.6 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
5.1 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
1.8 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
1.8 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.3 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 |
0.2 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
4.2 |
GO:0043524 |
negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 |
2.0 |
GO:2001243 |
negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 |
2.7 |
GO:0002230 |
positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 |
0.4 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
1.0 |
GO:0006970 |
response to osmotic stress(GO:0006970) |
| 0.0 |
0.3 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.4 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.1 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.0 |
0.3 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.0 |
0.1 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
1.6 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.0 |
0.5 |
GO:0006284 |
base-excision repair(GO:0006284) |