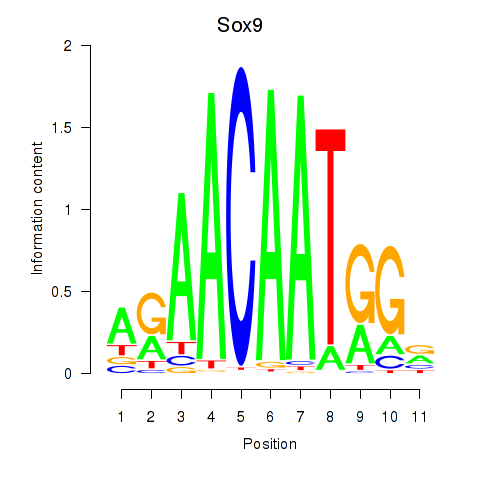
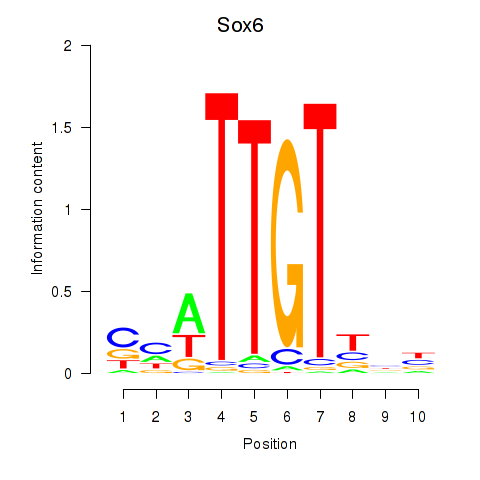
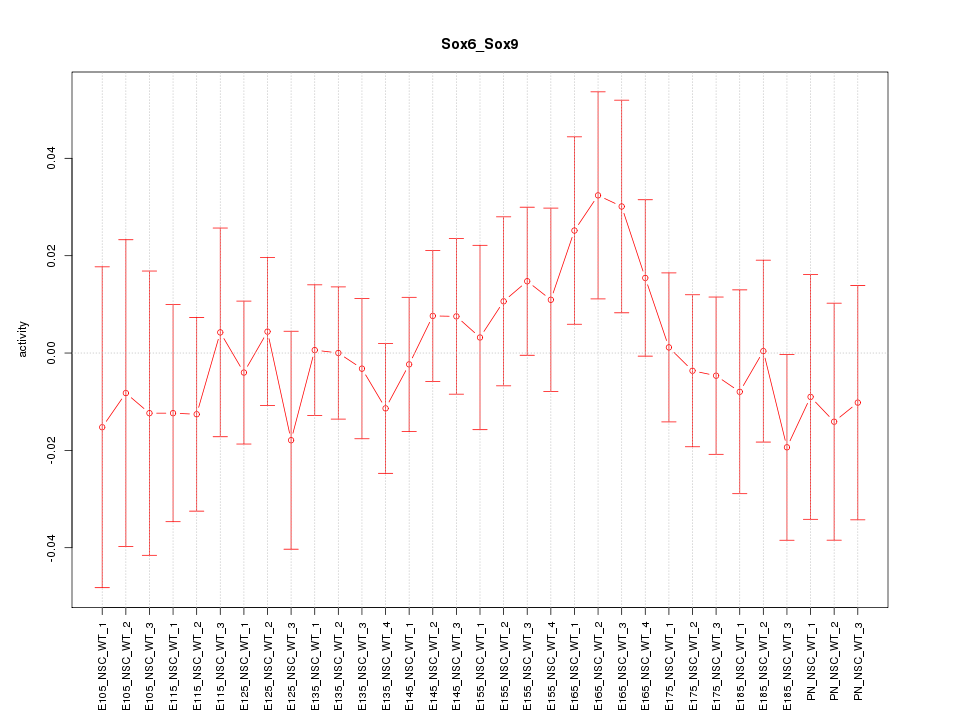
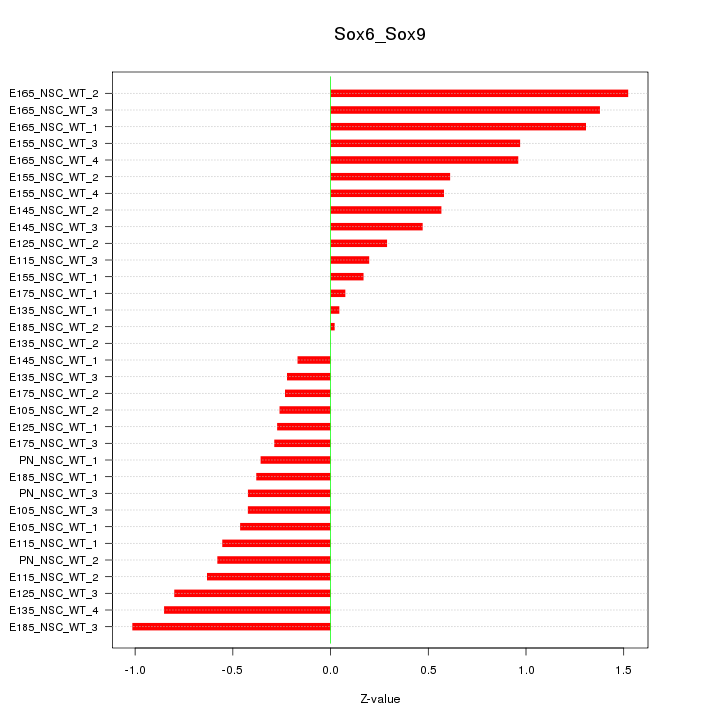
| 2.2 |
6.6 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.9 |
2.8 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.8 |
3.1 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.7 |
2.1 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.6 |
2.4 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.4 |
3.1 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.4 |
2.1 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.4 |
4.1 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.4 |
1.5 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 |
1.6 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 |
1.9 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 |
1.0 |
GO:0043379 |
memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.3 |
1.2 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.3 |
0.9 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.3 |
3.8 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 |
1.6 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.3 |
4.4 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.3 |
3.1 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 |
0.8 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 |
2.3 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 |
0.6 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 |
0.8 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 |
0.8 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.2 |
1.0 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.2 |
0.7 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 |
1.5 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 0.2 |
3.1 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.2 |
1.5 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 |
0.7 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.2 |
0.5 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 |
1.1 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 |
2.9 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 |
1.4 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.4 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 |
1.1 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
1.0 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.5 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
1.5 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.1 |
1.7 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.5 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.6 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 |
0.4 |
GO:1902524 |
positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 |
1.3 |
GO:0038180 |
nerve growth factor signaling pathway(GO:0038180) |
| 0.1 |
0.7 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.1 |
0.5 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.5 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 |
0.7 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.3 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.2 |
GO:2000790 |
regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.1 |
0.5 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
2.5 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
1.1 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
1.0 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.1 |
0.8 |
GO:0098907 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 |
1.2 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.3 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 |
0.3 |
GO:0090325 |
regulation of locomotion involved in locomotory behavior(GO:0090325) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.2 |
GO:0090285 |
regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 |
0.8 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.1 |
0.6 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
1.1 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.2 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.1 |
0.2 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 |
0.2 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 |
0.6 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
0.5 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 |
1.0 |
GO:0048505 |
regulation of timing of cell differentiation(GO:0048505) |
| 0.1 |
0.1 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.4 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.5 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 |
0.3 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
1.9 |
GO:0010043 |
response to zinc ion(GO:0010043) |
| 0.1 |
0.8 |
GO:0090179 |
planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 |
0.4 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 |
1.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.6 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.1 |
1.9 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.3 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 |
0.1 |
GO:0060700 |
regulation of ribonuclease activity(GO:0060700) |
| 0.1 |
0.3 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.1 |
1.7 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.3 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.2 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.1 |
GO:0060743 |
estrous cycle(GO:0044849) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 |
1.1 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.4 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.5 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
2.8 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.6 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.8 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.7 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.1 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.3 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.2 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.1 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.3 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.2 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 |
0.2 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
0.4 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 |
0.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
1.0 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 |
1.9 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.0 |
1.3 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.0 |
1.4 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.2 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.0 |
0.1 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
2.0 |
GO:0048814 |
regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 |
1.5 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.0 |
1.2 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.5 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.3 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.4 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.5 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.8 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.3 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.4 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
2.3 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.0 |
0.2 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.3 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.1 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 |
2.0 |
GO:0030705 |
cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 |
1.0 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
2.2 |
GO:0010506 |
regulation of autophagy(GO:0010506) |
| 0.0 |
0.3 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.6 |
GO:0030517 |
negative regulation of axon extension(GO:0030517) |
| 0.0 |
0.0 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 |
0.7 |
GO:0007018 |
microtubule-based movement(GO:0007018) |
| 0.0 |
0.1 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.1 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.2 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |