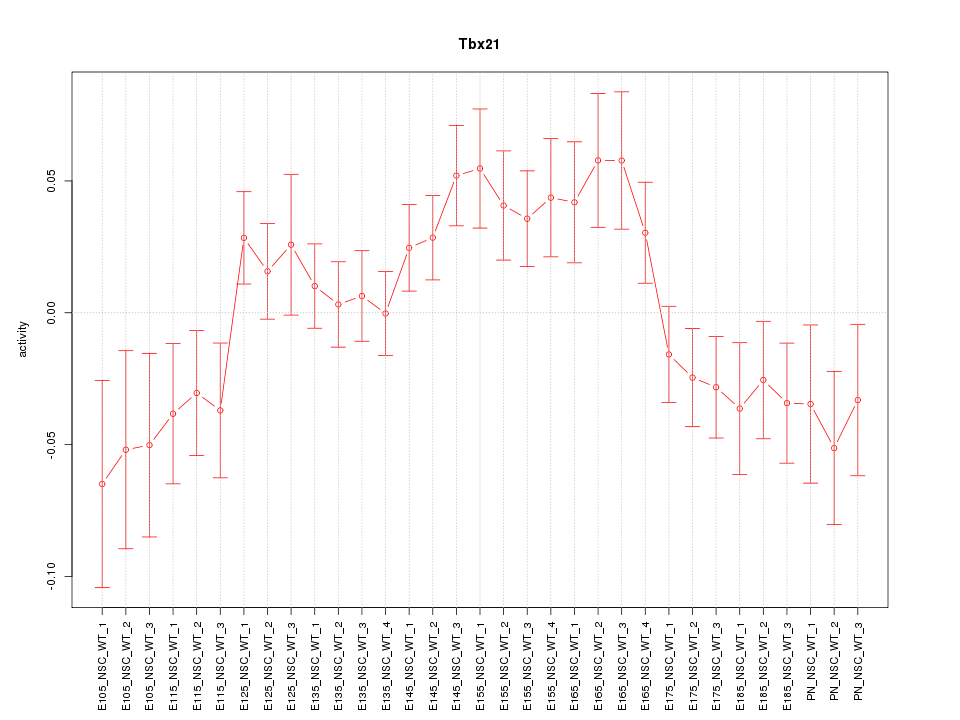
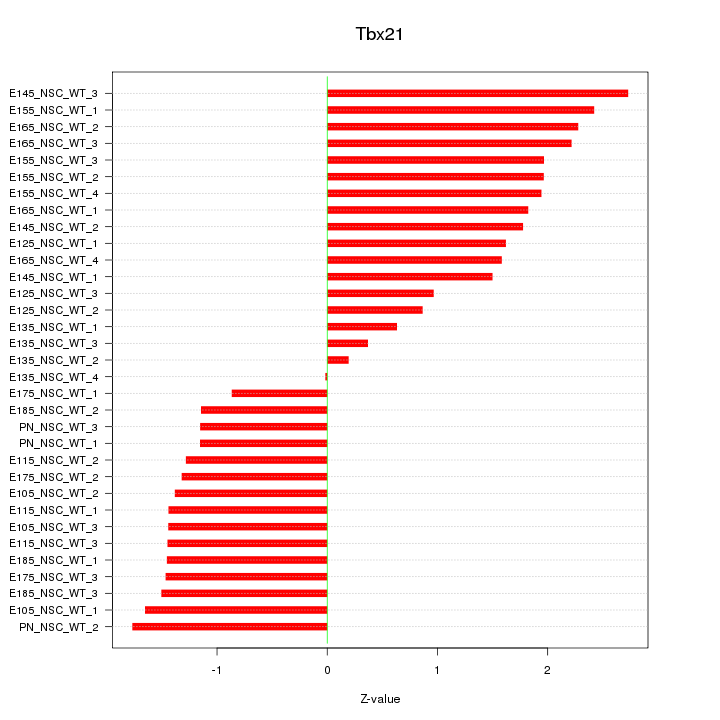
Motif ID: Tbx21
Z-value: 1.554
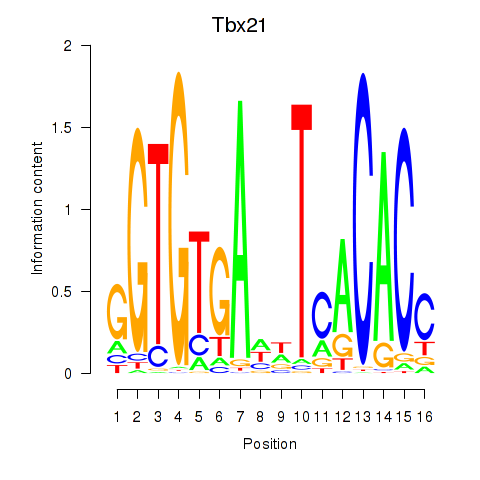
Transcription factors associated with Tbx21:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbx21 | ENSMUSG00000001444.2 | Tbx21 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0021649 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.4 | 22.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.0 | 2.9 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.9 | 3.7 | GO:0010040 | response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) cellular response to copper ion(GO:0071280) regulation of peroxidase activity(GO:2000468) |
| 0.8 | 5.0 | GO:0032796 | uropod organization(GO:0032796) |
| 0.8 | 2.5 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 0.8 | 2.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.7 | 2.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.7 | 2.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.6 | 8.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.6 | 9.0 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.5 | 4.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.4 | 18.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.4 | 4.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 1.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.4 | 2.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.4 | 1.2 | GO:0048296 | negative regulation of isotype switching(GO:0045829) isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.4 | 2.2 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 | 1.9 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.3 | 2.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 0.8 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.3 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.7 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.2 | 2.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 1.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 4.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 1.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 | 1.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.2 | 1.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 0.5 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.2 | 1.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 4.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 1.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 6.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 2.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.0 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 4.0 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 | 0.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 7.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 2.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.6 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.7 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.9 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 2.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 5.5 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 1.7 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 1.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.3 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 2.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.8 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 1.7 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.5 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.4 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.8 | 2.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 2.9 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 2.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 4.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 2.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 1.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 22.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 5.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 0.7 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.2 | 0.5 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 7.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 1.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 3.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 4.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 4.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 8.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 7.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.4 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.8 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.2 | 8.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 1.1 | 9.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.1 | 4.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 1.0 | 4.9 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.9 | 2.6 | GO:0050827 | toxin receptor binding(GO:0050827) |
| 0.7 | 2.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.7 | 2.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.6 | 3.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.5 | 5.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 2.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 2.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 1.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.4 | 1.6 | GO:0071532 | ornithine decarboxylase inhibitor activity(GO:0008073) ankyrin repeat binding(GO:0071532) |
| 0.4 | 1.9 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 4.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 4.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 5.5 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.2 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 3.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 6.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 4.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 2.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 2.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 16.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 3.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 2.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 8.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.4 | GO:0008905 | mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 25.6 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.4 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.0 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) DNA-methyltransferase activity(GO:0009008) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 6.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |