| 0.5 |
2.8 |
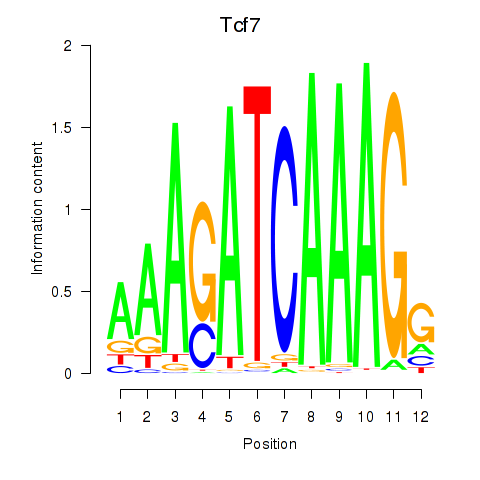
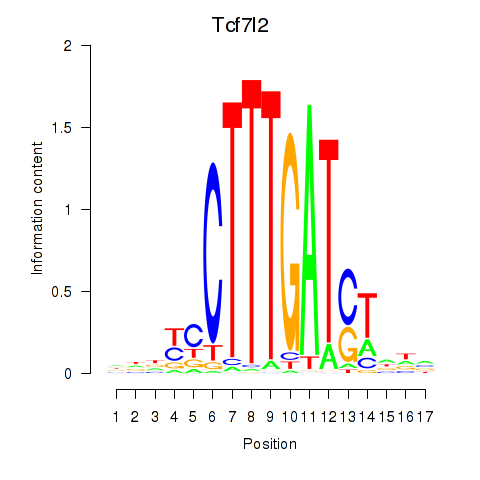
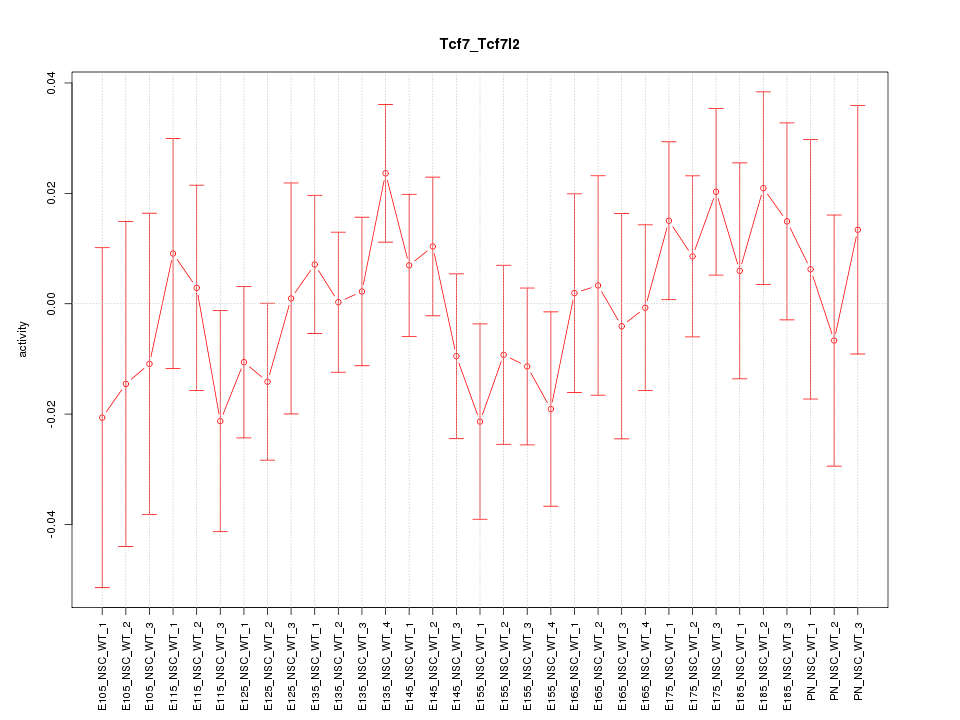
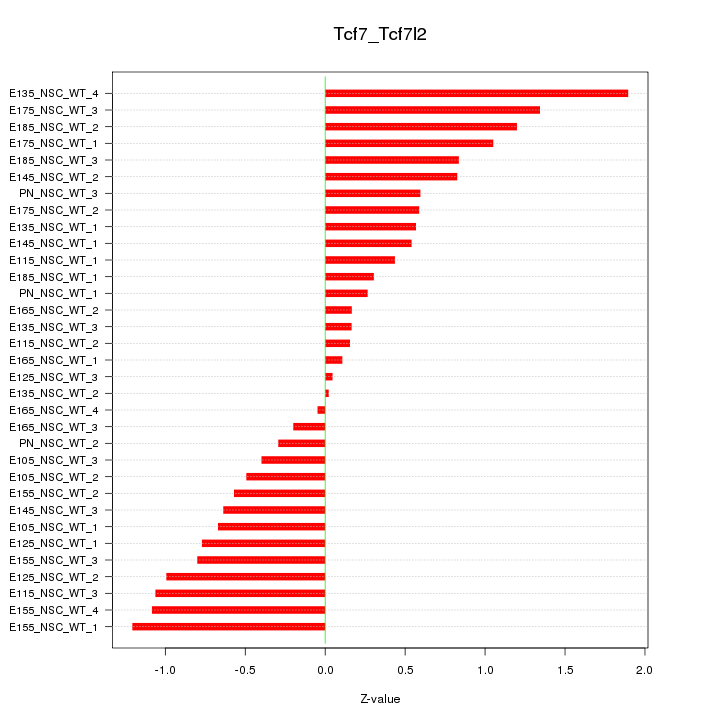
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.4 |
2.5 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 |
1.0 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.2 |
0.9 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 |
0.7 |
GO:0046104 |
thymidine metabolic process(GO:0046104) |
| 0.2 |
1.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.2 |
0.9 |
GO:2000054 |
negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 |
0.7 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 |
0.5 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.2 |
0.8 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 |
0.8 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.1 |
0.4 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 |
0.4 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 |
0.6 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 |
0.9 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
0.8 |
GO:0061365 |
positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 |
0.4 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 |
0.9 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.6 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.1 |
0.6 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 |
0.3 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
0.5 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
0.4 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 |
0.7 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 |
0.3 |
GO:0042939 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 |
0.3 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.1 |
1.1 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
0.7 |
GO:0036376 |
sodium ion export from cell(GO:0036376) |
| 0.1 |
0.2 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.1 |
0.3 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.6 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) |
| 0.1 |
0.5 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
1.0 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
0.3 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 |
0.4 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 |
0.2 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.1 |
0.2 |
GO:2000813 |
actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 |
1.1 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 |
0.9 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.3 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.0 |
0.9 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.6 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
1.6 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.5 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.1 |
GO:0060468 |
prevention of polyspermy(GO:0060468) |
| 0.0 |
1.0 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.2 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.0 |
0.6 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 |
0.3 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.3 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 |
0.5 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.0 |
0.3 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.2 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.0 |
0.2 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 |
0.5 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 |
0.3 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 |
0.1 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 |
0.1 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.0 |
0.3 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 |
1.1 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.1 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
2.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.3 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 |
0.3 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.2 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.0 |
0.1 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.0 |
0.5 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.0 |
0.2 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
0.4 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 |
0.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
0.4 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.5 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.2 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.0 |
0.5 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.5 |
GO:2000249 |
regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 |
0.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.8 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
2.1 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.1 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.2 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.2 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.0 |
1.2 |
GO:0018107 |
peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 |
0.7 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.9 |
GO:0035383 |
acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 |
0.1 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.7 |
GO:0001756 |
somitogenesis(GO:0001756) |
| 0.0 |
0.7 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
1.9 |
GO:0006275 |
regulation of DNA replication(GO:0006275) |
| 0.0 |
0.4 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.1 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |