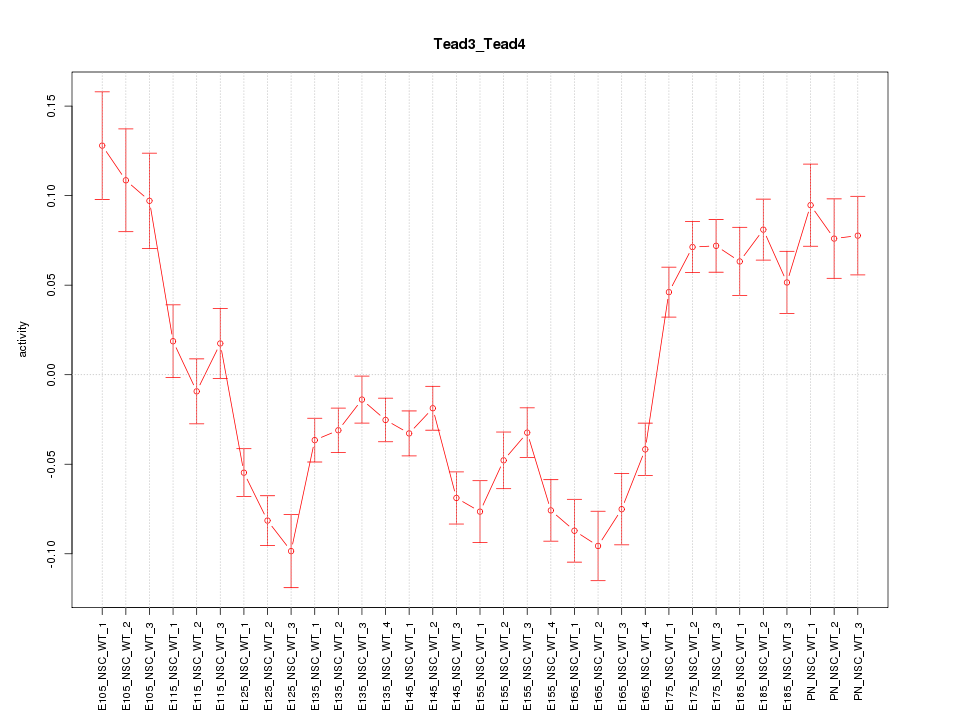
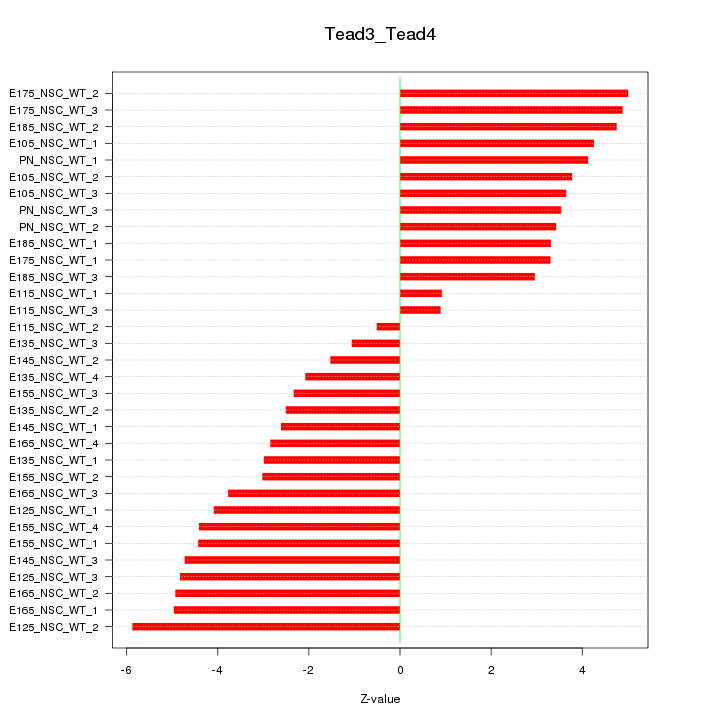
Motif ID: Tead3_Tead4
Z-value: 3.664
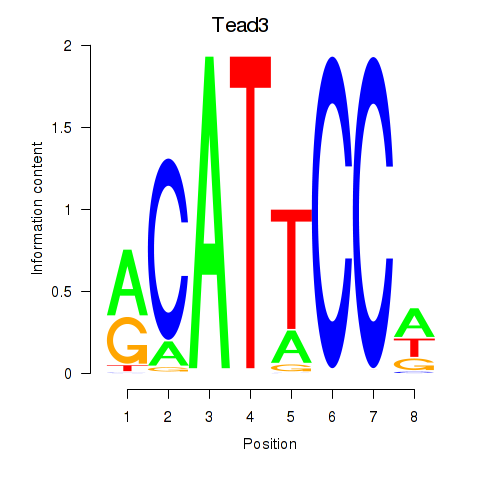
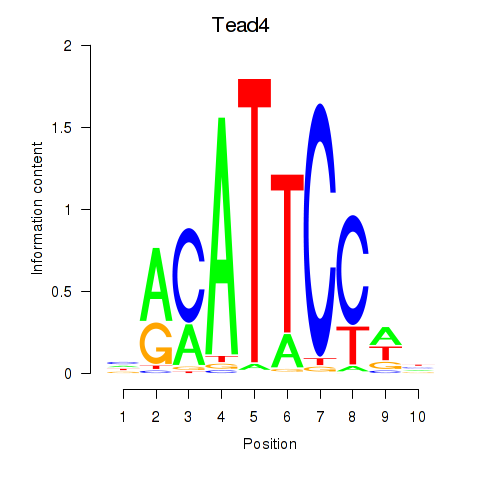
Transcription factors associated with Tead3_Tead4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tead3 | ENSMUSG00000002249.11 | Tead3 |
| Tead4 | ENSMUSG00000030353.9 | Tead4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead3 | mm10_v2_chr17_-_28350600_28350681 | 0.39 | 2.3e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.8 | 79.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 9.5 | 28.5 | GO:2001139 | negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 9.2 | 46.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 5.0 | 20.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 4.1 | 16.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 4.0 | 19.9 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 4.0 | 11.9 | GO:0044849 | estrous cycle(GO:0044849) |
| 3.9 | 11.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 3.8 | 30.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 3.7 | 25.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 3.6 | 21.4 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 3.5 | 7.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 3.2 | 15.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 3.1 | 12.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 2.9 | 5.8 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 2.8 | 8.4 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 2.8 | 8.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 2.7 | 2.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 2.6 | 13.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 2.6 | 31.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 2.4 | 7.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) retinal rod cell differentiation(GO:0060221) renal vesicle induction(GO:0072034) |
| 2.0 | 7.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.9 | 7.6 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 1.9 | 42.6 | GO:0001946 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 1.7 | 8.7 | GO:1903598 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) |
| 1.7 | 5.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.6 | 4.9 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 1.6 | 11.3 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 1.6 | 4.7 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 1.5 | 10.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 1.5 | 6.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.5 | 10.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.5 | 4.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.5 | 4.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.4 | 8.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.4 | 5.6 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.4 | 8.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 1.4 | 2.7 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 1.3 | 4.0 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 1.3 | 6.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.3 | 3.8 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 1.2 | 17.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 1.2 | 10.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 1.1 | 4.6 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 1.1 | 9.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 1.1 | 12.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 1.1 | 3.3 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 1.0 | 9.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.0 | 3.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 1.0 | 2.9 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.9 | 15.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.9 | 3.8 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.9 | 3.7 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.9 | 5.5 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.9 | 7.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.9 | 10.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 | 7.9 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.8 | 4.2 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.8 | 6.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.8 | 5.8 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.8 | 2.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.8 | 8.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.8 | 3.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.8 | 3.8 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.7 | 3.7 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.7 | 0.7 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.7 | 2.9 | GO:1903416 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 0.7 | 5.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.6 | 6.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.6 | 1.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.6 | 3.1 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.6 | 6.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.6 | 11.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.6 | 4.6 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.6 | 6.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.6 | 5.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.6 | 2.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.6 | 7.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.5 | 1.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.5 | 10.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.5 | 6.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.5 | 5.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.5 | 3.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.5 | 24.7 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.5 | 1.0 | GO:0045608 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.5 | 7.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 1.8 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.4 | 10.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 1.3 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.4 | 3.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.4 | 6.3 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.4 | 2.9 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.4 | 2.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.4 | 1.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 5.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 1.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.4 | 2.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 2.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.4 | 12.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.4 | 4.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 2.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 3.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 13.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.3 | 4.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.3 | 3.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 2.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 4.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 4.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 6.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.3 | 2.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 2.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 1.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 2.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 8.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.3 | 4.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 1.0 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 1.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.3 | 0.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.3 | 1.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 0.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 15.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 2.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 1.2 | GO:0071205 | clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 | 6.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 3.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 2.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 1.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.8 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 8.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 2.8 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 2.1 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 6.2 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.2 | 11.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.2 | 5.4 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.2 | 11.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.2 | 2.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 2.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 2.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 1.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.2 | 0.5 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.2 | 1.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 8.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.2 | 4.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 2.0 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.1 | 15.5 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 0.7 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 5.8 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 0.5 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 | 9.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.7 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 2.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 2.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 1.8 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.2 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 1.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.2 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 1.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 2.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 2.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 2.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 3.9 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.1 | 1.4 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 1.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 10.5 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.1 | 1.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 3.8 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.9 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 2.2 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.1 | 2.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 4.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.8 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.8 | GO:0003229 | ventricular cardiac muscle tissue development(GO:0003229) |
| 0.0 | 2.3 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.8 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 1.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 3.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.2 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 1.5 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.3 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.4 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.4 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 46.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 9.5 | 28.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 7.1 | 21.4 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 6.8 | 20.4 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 3.9 | 11.8 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 2.5 | 10.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 2.2 | 36.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.8 | 12.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.7 | 25.8 | GO:0043196 | varicosity(GO:0043196) |
| 1.5 | 12.4 | GO:0098536 | deuterosome(GO:0098536) |
| 1.5 | 77.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 1.4 | 9.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.3 | 18.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 3.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.0 | 4.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.0 | 8.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.9 | 3.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.9 | 2.8 | GO:0034679 | integrin alpha2-beta1 complex(GO:0034666) integrin alpha3-beta1 complex(GO:0034667) integrin alpha9-beta1 complex(GO:0034679) |
| 0.9 | 11.9 | GO:0043219 | lateral loop(GO:0043219) |
| 0.9 | 5.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 3.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.7 | 2.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.7 | 5.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 7.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.6 | 6.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 5.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.5 | 2.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.5 | 7.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 5.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.5 | 8.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.5 | 5.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 7.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.5 | 2.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 9.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 21.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 5.8 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.4 | 3.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 1.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.4 | 3.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 4.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 36.8 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 4.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 1.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 1.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 7.2 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 5.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 3.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 10.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.3 | 1.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 14.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 6.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 90.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.2 | 24.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 2.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 9.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 7.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 12.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 8.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 10.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 4.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 14.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 4.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 12.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 20.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 2.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 6.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 17.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 5.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 9.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 5.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 6.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 8.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 5.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 4.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 7.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 46.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 44.8 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 5.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 6.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 28.5 | GO:0046911 | hydroxyapatite binding(GO:0046848) metal chelating activity(GO:0046911) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 5.3 | 21.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 5.1 | 46.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 4.4 | 97.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 4.4 | 8.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 3.9 | 11.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 3.6 | 17.9 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 2.7 | 10.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 2.4 | 11.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 2.4 | 23.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 2.4 | 7.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 2.3 | 18.2 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 2.2 | 13.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 2.1 | 25.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 2.0 | 10.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.8 | 11.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.7 | 8.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.7 | 6.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.6 | 35.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 1.3 | 4.0 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 1.3 | 5.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.0 | 4.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.0 | 8.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.0 | 3.8 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.9 | 2.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.9 | 4.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.9 | 10.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 2.7 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.9 | 6.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.8 | 3.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.8 | 6.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.8 | 2.4 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.8 | 1.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.8 | 46.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.8 | 5.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.8 | 2.3 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.7 | 8.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.7 | 2.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.7 | 8.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.7 | 13.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.7 | 2.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.7 | 10.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.7 | 24.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.7 | 6.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.6 | 7.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.6 | 8.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.6 | 3.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.6 | 2.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 15.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 12.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.5 | 4.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.5 | 2.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.5 | 7.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 1.8 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 14.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 4.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 4.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 17.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.4 | 2.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 4.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 8.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 5.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 4.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.4 | 11.8 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.3 | 3.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 9.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 1.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 1.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.3 | 5.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.3 | 12.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 10.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 3.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 5.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 38.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 3.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 2.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 10.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 0.8 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.3 | 4.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 2.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 18.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 8.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 14.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 6.0 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.2 | 2.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 28.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 14.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.2 | 3.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 1.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 18.4 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.2 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 2.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 2.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 3.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 10.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 5.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 6.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 5.8 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 2.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 2.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 3.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.4 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 3.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 20.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 3.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 6.0 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 3.9 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 10.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 2.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.6 | GO:0017161 | JUN kinase phosphatase activity(GO:0008579) phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 1.6 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 4.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 5.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |