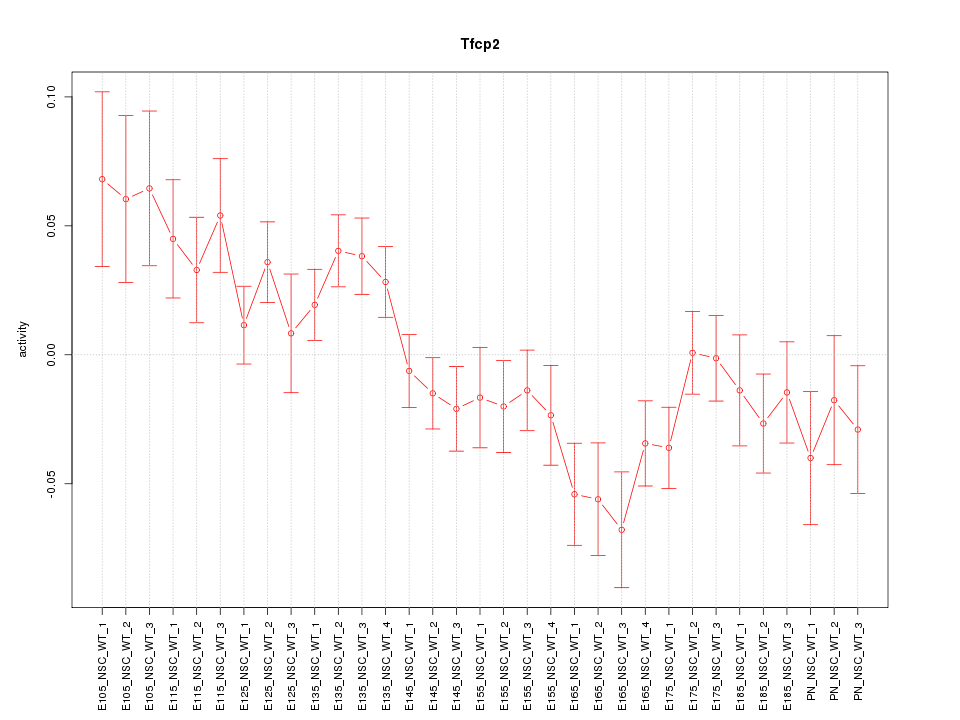
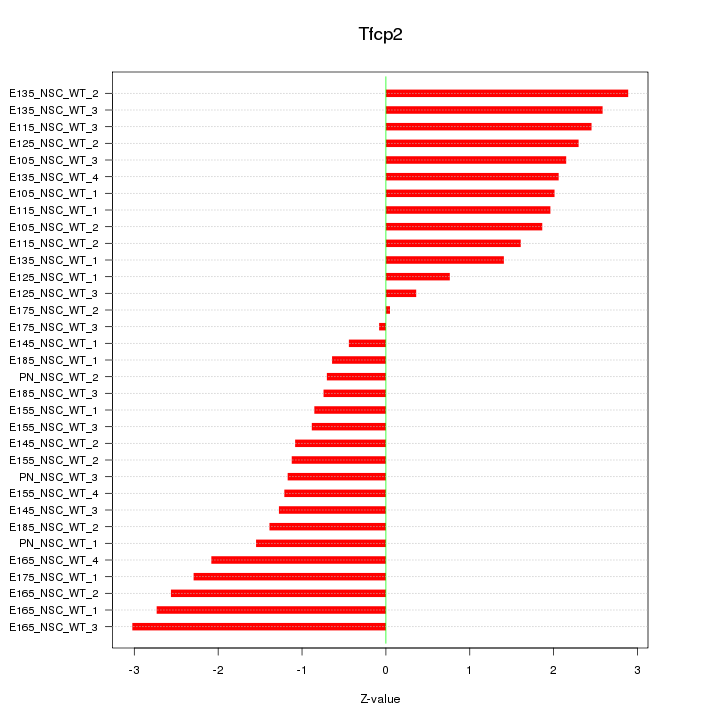
Motif ID: Tfcp2
Z-value: 1.732
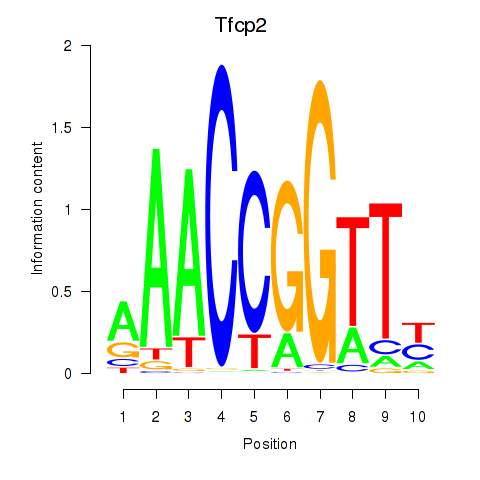
Transcription factors associated with Tfcp2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfcp2 | ENSMUSG00000009733.8 | Tfcp2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2 | mm10_v2_chr15_-_100551959_100552010 | -0.56 | 7.0e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.5 | 4.6 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.5 | 7.3 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 1.4 | 4.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.3 | 4.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 1.3 | 5.2 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 1.3 | 6.3 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 1.1 | 11.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 1.1 | 3.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 1.1 | 3.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.9 | 3.5 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.9 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.9 | 4.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.8 | 2.4 | GO:0045950 | female meiosis chromosome segregation(GO:0016321) negative regulation of mitotic recombination(GO:0045950) |
| 0.8 | 3.2 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.8 | 2.3 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.7 | 4.5 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.7 | 2.2 | GO:0046078 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.7 | 3.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.7 | 2.8 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.7 | 3.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.7 | 4.7 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.7 | 10.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.7 | 2.6 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.6 | 5.9 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.6 | 7.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.5 | 4.4 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.5 | 3.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.5 | 2.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.5 | 3.4 | GO:2000781 | DNA dealkylation involved in DNA repair(GO:0006307) positive regulation of double-strand break repair(GO:2000781) |
| 0.5 | 1.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 2.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.4 | 2.6 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.4 | 1.7 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.4 | 8.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.4 | 12.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.4 | 2.1 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.3 | 1.0 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.3 | 1.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 4.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 1.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 2.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 2.2 | GO:0032782 | bile acid secretion(GO:0032782) positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.3 | 5.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 1.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 3.6 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.3 | 2.5 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.3 | 1.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.2 | 2.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 3.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 0.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 1.6 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 1.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 1.9 | GO:0031424 | keratinization(GO:0031424) establishment of skin barrier(GO:0061436) |
| 0.2 | 1.0 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) |
| 0.2 | 1.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.2 | 1.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.8 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.2 | 1.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 0.6 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 2.6 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 6.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 1.6 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.2 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 9.0 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.2 | 6.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 1.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.2 | 0.9 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 3.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 4.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 3.2 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 1.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 3.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.4 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 3.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 1.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 5.8 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 0.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 2.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 1.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 2.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 2.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.2 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.1 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 1.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.9 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 1.8 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 1.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 1.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.2 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 2.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 3.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 6.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.8 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 2.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 3.2 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 1.2 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.6 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.0 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 2.1 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0042659 | ventral spinal cord interneuron differentiation(GO:0021514) regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.6 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.3 | 6.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 1.1 | 9.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.0 | 6.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.9 | 3.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.9 | 3.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.8 | 3.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.8 | 4.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.8 | 3.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.7 | 2.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.6 | 2.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.5 | 3.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.5 | 2.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.5 | 4.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.4 | 4.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 2.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 5.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 1.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 10.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 2.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 2.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 3.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 4.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 2.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 6.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 7.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 2.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 4.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 9.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 3.6 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 15.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 5.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.7 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.0 | 5.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.5 | 4.6 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.3 | 6.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 1.2 | 3.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.9 | 2.8 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.9 | 2.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.8 | 3.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.8 | 2.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.8 | 4.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.8 | 9.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.7 | 2.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.7 | 3.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.6 | 2.5 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.6 | 4.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 3.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.5 | 2.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.5 | 1.5 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.5 | 2.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.5 | 1.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 1.8 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 10.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 3.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.4 | 4.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 3.0 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.4 | 3.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 3.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 1.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 4.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.7 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.3 | 1.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 2.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 8.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.2 | 6.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 3.0 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.2 | 4.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 2.6 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.2 | 1.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 1.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 6.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 7.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 2.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 4.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 1.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 2.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 3.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.7 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.1 | 1.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 17.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 3.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 2.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.6 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 3.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 7.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 1.8 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 1.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 7.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 2.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 4.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 4.9 | GO:0003682 | chromatin binding(GO:0003682) |