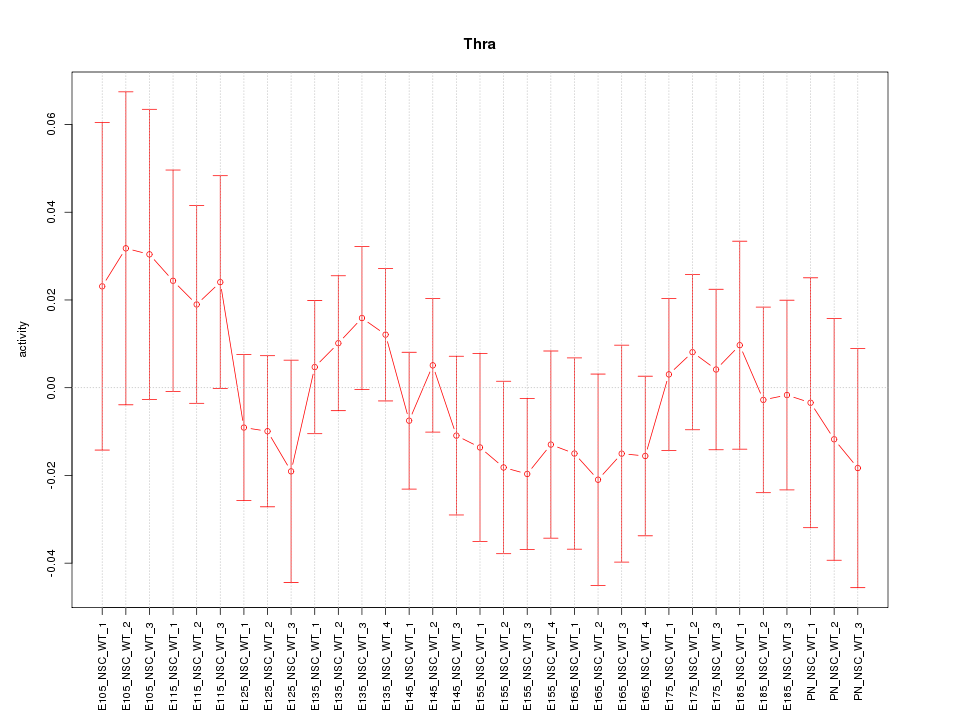
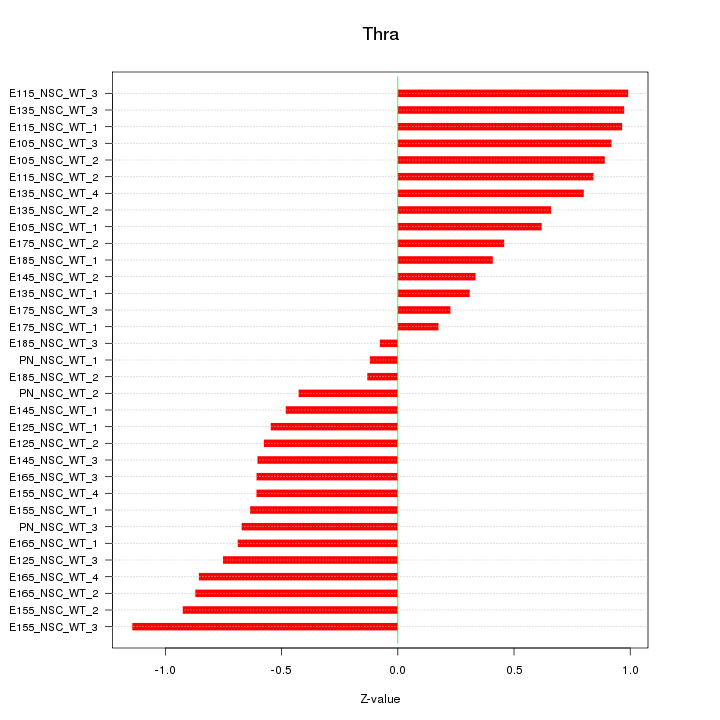
| 0.7 |
2.0 |
GO:0090202 |
regulation of primitive erythrocyte differentiation(GO:0010725) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.6 |
1.9 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 0.6 |
2.4 |
GO:0098763 |
mitotic cell cycle phase(GO:0098763) |
| 0.5 |
1.5 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.5 |
1.5 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.4 |
4.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 |
2.1 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.4 |
1.6 |
GO:0051295 |
establishment of meiotic spindle localization(GO:0051295) |
| 0.4 |
1.6 |
GO:0071105 |
response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.4 |
1.1 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.4 |
1.1 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.3 |
2.8 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.3 |
4.0 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 |
1.3 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 |
0.8 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 |
3.4 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.2 |
1.0 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.2 |
1.6 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.2 |
1.9 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 0.2 |
1.3 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 |
1.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 |
1.3 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.2 |
1.4 |
GO:0061084 |
negative regulation of protein refolding(GO:0061084) |
| 0.2 |
1.0 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.2 |
0.5 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 |
1.4 |
GO:0019240 |
citrulline biosynthetic process(GO:0019240) |
| 0.2 |
0.8 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) |
| 0.1 |
0.4 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) blood vessel lumenization(GO:0072554) regulation of heart induction(GO:0090381) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 |
1.5 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 |
0.7 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 |
1.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.9 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.7 |
GO:0035989 |
tendon development(GO:0035989) |
| 0.1 |
0.6 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 |
0.7 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 |
0.6 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.1 |
1.2 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
3.5 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 |
0.9 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 |
0.2 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.1 |
1.3 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.6 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 |
0.4 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) enamel mineralization(GO:0070166) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 |
1.2 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 |
0.5 |
GO:1903753 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) hard palate development(GO:0060022) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 |
0.7 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.1 |
0.6 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.1 |
1.5 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 |
2.0 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.1 |
0.4 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 |
2.7 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.1 |
1.4 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.1 |
0.4 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 |
1.2 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
2.6 |
GO:0043297 |
apical junction assembly(GO:0043297) |
| 0.1 |
0.3 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.8 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.3 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.3 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.0 |
0.8 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.3 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
1.3 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.5 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
3.2 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
3.7 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.2 |
GO:0051004 |
plasma membrane to endosome transport(GO:0048227) regulation of lipoprotein lipase activity(GO:0051004) |
| 0.0 |
0.3 |
GO:0003214 |
cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 |
0.3 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.6 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.8 |
GO:1900027 |
regulation of ruffle assembly(GO:1900027) |
| 0.0 |
0.1 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 |
0.7 |
GO:1902230 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 |
0.1 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
0.4 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.1 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 |
0.3 |
GO:0002385 |
mucosal immune response(GO:0002385) |
| 0.0 |
0.2 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.6 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.9 |
GO:0045840 |
positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 |
1.0 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.7 |
GO:0031111 |
negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 |
0.4 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.4 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
1.6 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.0 |
0.1 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 |
0.1 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
1.6 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.9 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.2 |
GO:1901673 |
regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 |
0.1 |
GO:0060979 |
vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 |
0.2 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
2.8 |
GO:0060348 |
bone development(GO:0060348) |
| 0.0 |
0.6 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.2 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.1 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.7 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.4 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.3 |
GO:1902108 |
regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 |
0.3 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.1 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.0 |
0.1 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |