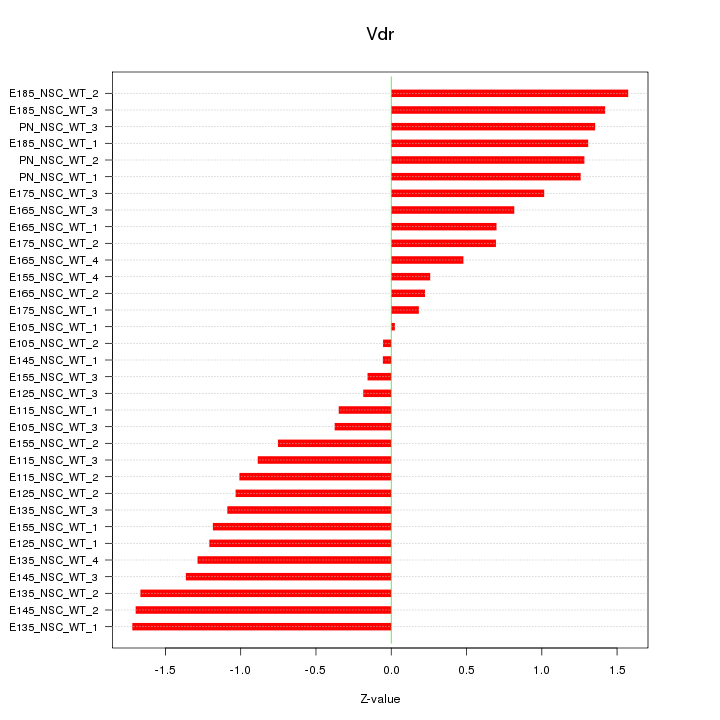
Motif ID: Vdr
Z-value: 1.019
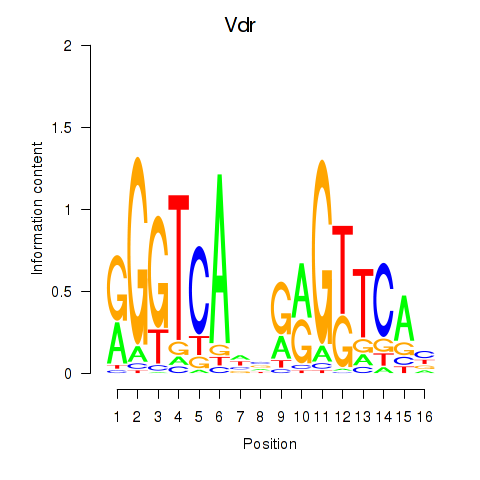
Transcription factors associated with Vdr:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Vdr | ENSMUSG00000022479.9 | Vdr |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 1.8 | 7.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 1.1 | 4.4 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.9 | 2.8 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.9 | 6.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.8 | 2.4 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.7 | 4.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.7 | 2.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.7 | 2.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.6 | 3.9 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.5 | 5.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.5 | 1.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.4 | 7.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.4 | 1.2 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid catabolic process(GO:0019478) |
| 0.4 | 1.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.4 | 1.9 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.3 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 0.9 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.3 | 0.9 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 1.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 2.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 1.9 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.3 | 1.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 3.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.2 | 0.6 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 1.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 1.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 2.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 4.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.4 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.7 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 1.6 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 1.8 | GO:0090051 | angiogenesis involved in wound healing(GO:0060055) negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.8 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.3 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 5.6 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.5 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 2.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 1.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:1901896 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.8 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.4 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 1.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.8 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 1.7 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.9 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0030168 | platelet activation(GO:0030168) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 2.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 2.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 2.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 6.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 7.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 4.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 7.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 6.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 4.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 2.0 | 6.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.0 | 5.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.9 | 4.4 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.6 | 3.9 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.5 | 2.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 1.9 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.3 | 6.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 2.7 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.3 | 1.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) aspartate oxidase activity(GO:0015922) |
| 0.3 | 0.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 1.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 2.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.2 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.2 | 1.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 6.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 6.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.5 | GO:0019215 | phosphatidylinositol-3-phosphatase activity(GO:0004438) intermediate filament binding(GO:0019215) |
| 0.1 | 0.9 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.4 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.1 | 0.3 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 1.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.3 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 2.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 2.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0046933 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0015485 | cholesterol binding(GO:0015485) |