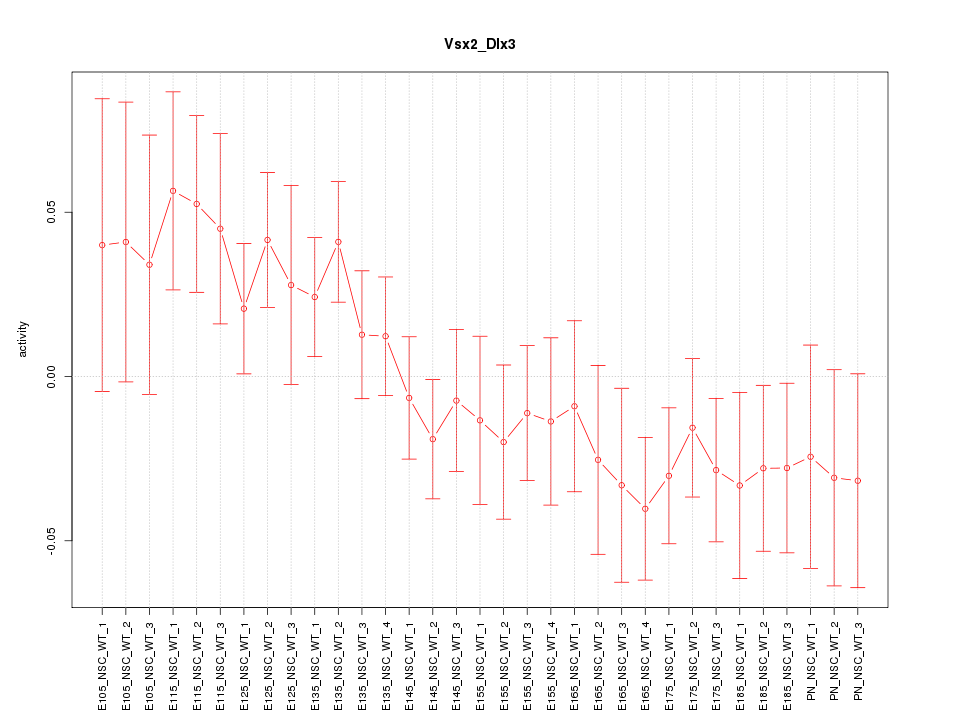
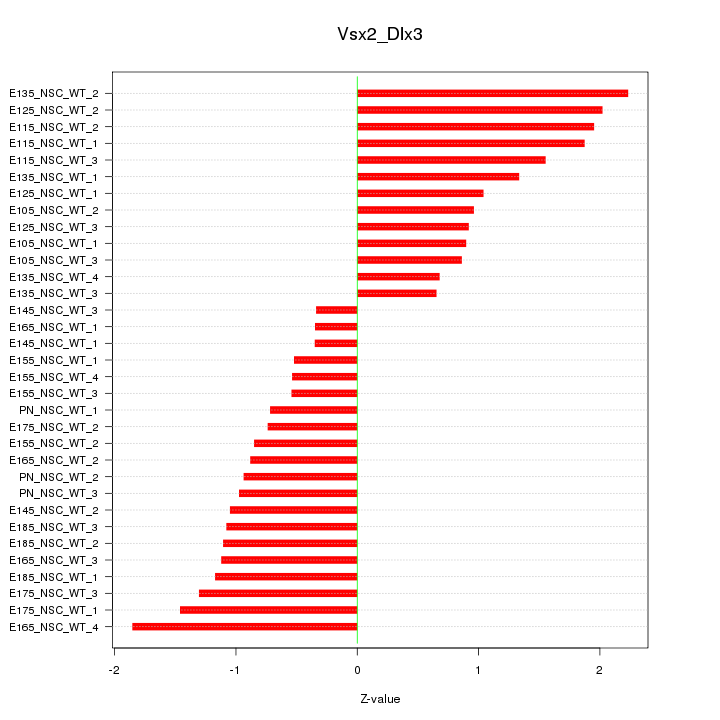
Motif ID: Vsx2_Dlx3
Z-value: 1.166
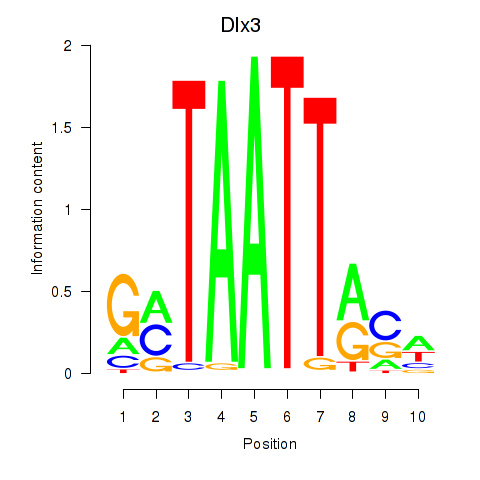
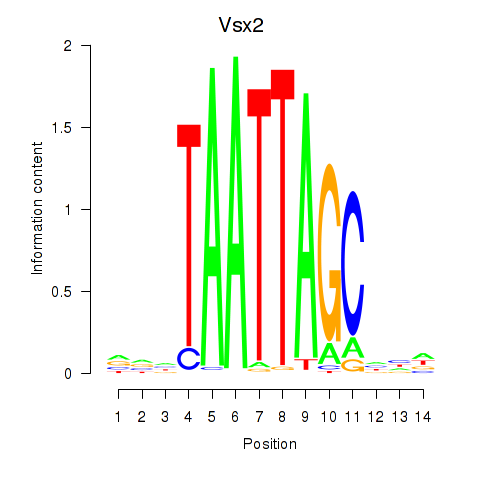
Transcription factors associated with Vsx2_Dlx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dlx3 | ENSMUSG00000001510.7 | Dlx3 |
| Vsx2 | ENSMUSG00000021239.6 | Vsx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vsx2 | mm10_v2_chr12_+_84569762_84569840 | 0.35 | 4.5e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 27.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 3.7 | 29.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 2.3 | 6.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.5 | 6.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.5 | 26.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.8 | 2.4 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.7 | 0.7 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.7 | 2.0 | GO:0090526 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.6 | 1.9 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.6 | 3.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.5 | 6.5 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.4 | 3.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.4 | 6.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 1.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.4 | 1.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.3 | 1.7 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 1.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 4.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.3 | 4.5 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.3 | 0.8 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.7 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 0.7 | GO:0060489 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 3.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 3.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 0.5 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 0.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.0 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.7 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 2.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.4 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 2.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 10.7 | GO:0008585 | female gonad development(GO:0008585) |
| 0.1 | 2.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 2.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 3.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.9 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.9 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 | 0.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 2.1 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 1.7 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 1.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.4 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0045333 | cellular respiration(GO:0045333) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 1.7 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 1.7 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.2 | 1.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.2 | 0.9 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 26.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 0.7 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 2.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 6.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 2.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.9 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 1.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 4.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.7 | 6.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.6 | 6.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.6 | 3.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 2.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 1.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 5.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 0.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 2.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 2.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 0.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 1.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.7 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 2.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 20.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 68.9 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.1 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 5.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.4 | GO:0000287 | magnesium ion binding(GO:0000287) |