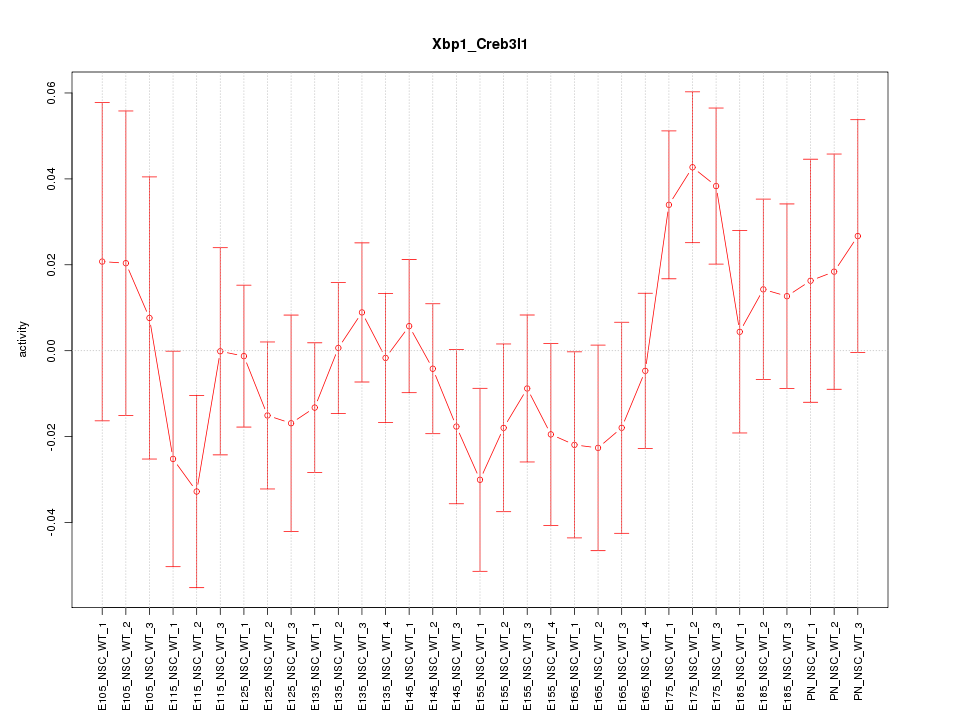
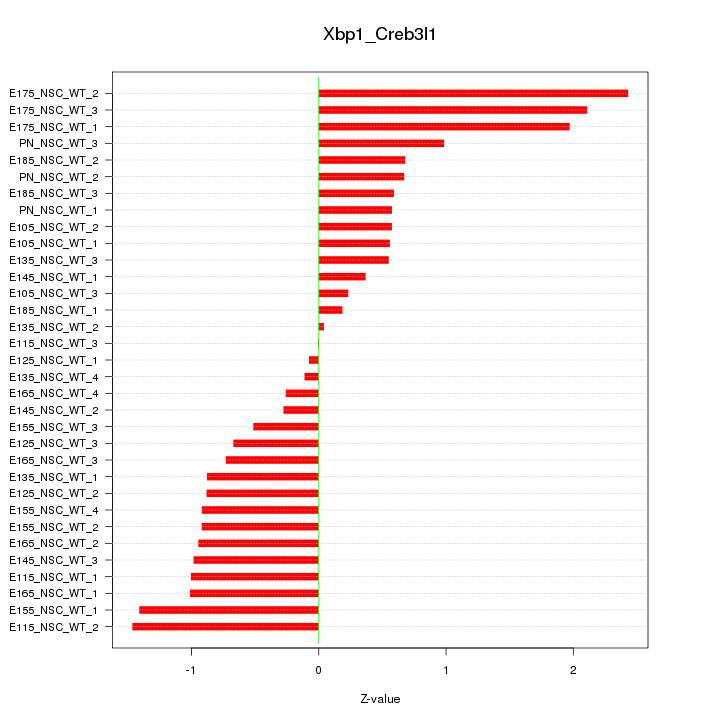
Motif ID: Xbp1_Creb3l1
Z-value: 0.964
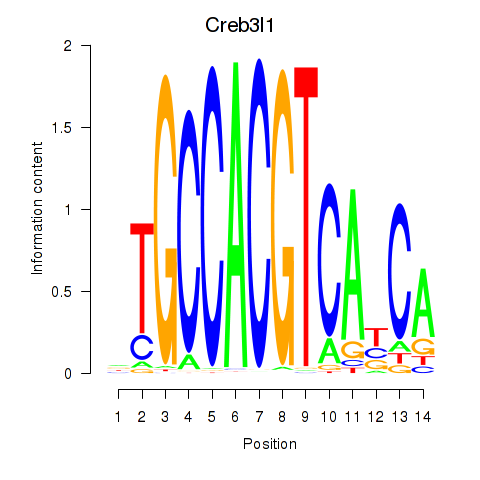
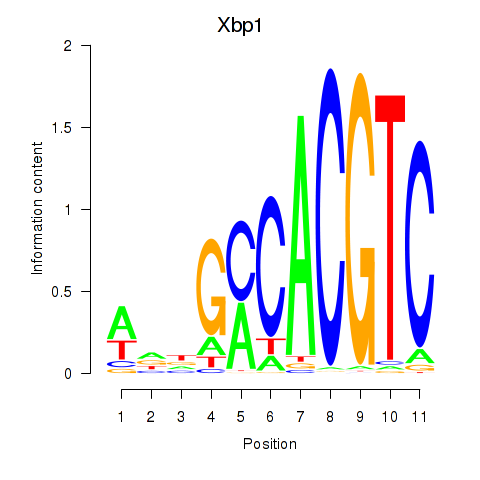
Transcription factors associated with Xbp1_Creb3l1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Creb3l1 | ENSMUSG00000027230.9 | Creb3l1 |
| Xbp1 | ENSMUSG00000020484.12 | Xbp1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l1 | mm10_v2_chr2_-_92024502_92024529 | 0.57 | 6.0e-04 | Click! |
| Xbp1 | mm10_v2_chr11_+_5520652_5520659 | -0.08 | 6.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 1.6 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.3 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 1.5 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.3 | 3.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.3 | 3.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.3 | 1.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.3 | 1.0 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.2 | 1.5 | GO:0032596 | protein transport into membrane raft(GO:0032596) dsRNA transport(GO:0033227) |
| 0.2 | 2.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.2 | 2.4 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 3.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 1.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.4 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 1.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 1.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.8 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.5 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 2.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 1.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.0 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.1 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.5 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.1 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.7 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.0 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 1.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.2 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 | 1.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 1.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.4 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.9 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.3 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 2.0 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 5.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 2.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 3.4 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 2.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 32.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 1.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 2.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 1.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 1.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.4 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.2 | 0.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.2 | 0.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.1 | 2.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.4 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.0 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 3.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.4 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008521 | acetyl-CoA transporter activity(GO:0008521) |
| 0.0 | 0.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 1.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 16.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 1.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |