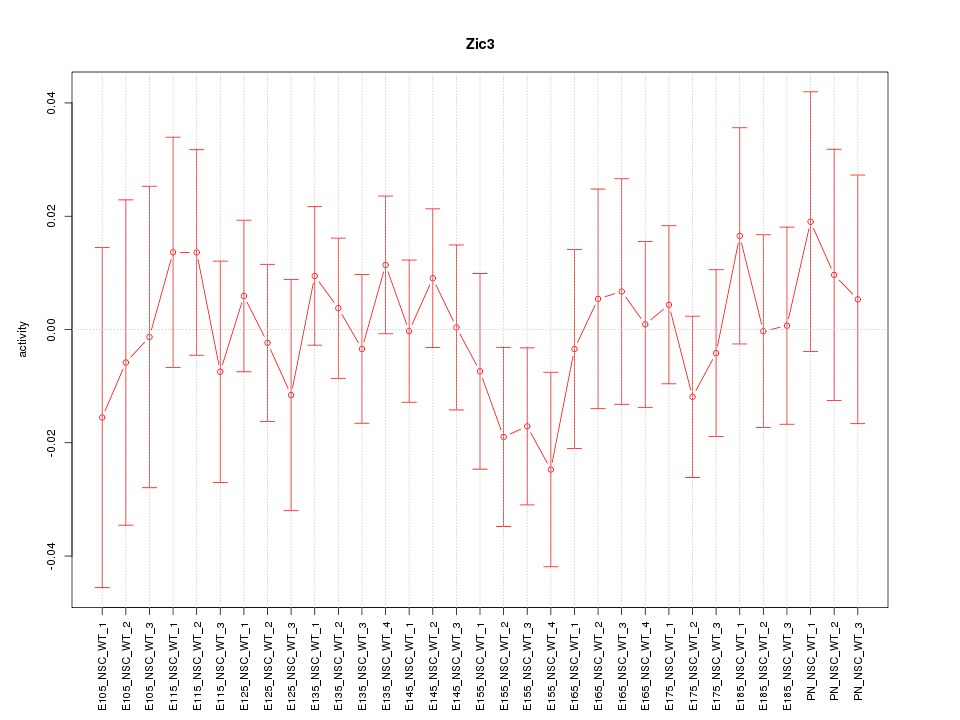
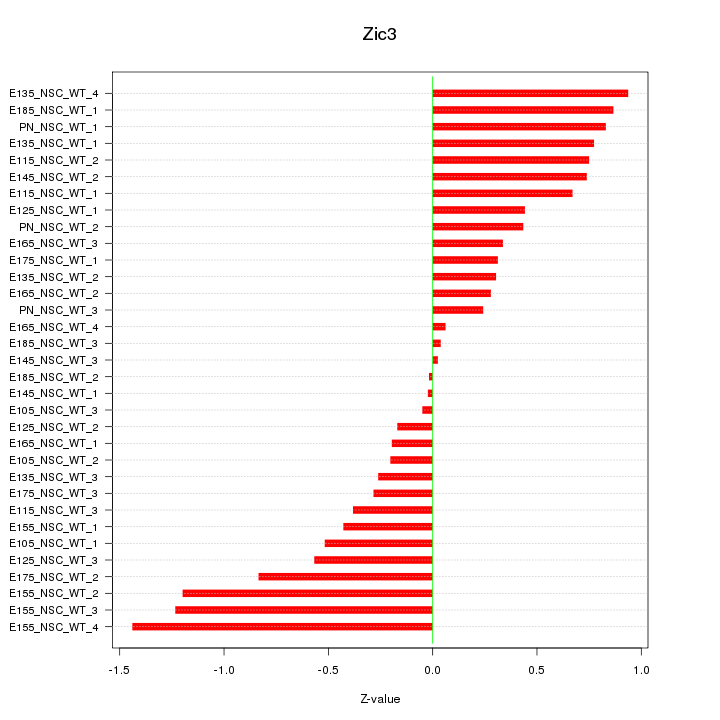
Motif ID: Zic3
Z-value: 0.608

Transcription factors associated with Zic3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zic3 | ENSMUSG00000067860.5 | Zic3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic3 | mm10_v2_chrX_+_58030622_58030643 | 0.24 | 1.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 1.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.3 | 1.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 1.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.2 | 0.5 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 | 0.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) glutamate secretion, neurotransmission(GO:0061535) |
| 0.2 | 0.9 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 0.6 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.2 | 0.6 | GO:0046671 | negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.2 | 1.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.7 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 1.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.3 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.9 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 0.3 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.1 | 0.3 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.8 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.4 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.1 | 0.7 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.1 | 0.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.1 | 0.2 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.5 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.1 | 0.7 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:1903598 | negative regulation of pinocytosis(GO:0048550) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:1902669 | positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) testosterone biosynthetic process(GO:0061370) renal vesicle induction(GO:0072034) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.1 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.3 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.1 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.5 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.3 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0090292 | nuclear migration along microfilament(GO:0031022) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 0.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.9 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.1 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 0.4 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 1.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 0.6 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 1.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 0.9 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.9 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 0.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 1.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.0 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |