Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for bbx
Z-value: 1.34
Transcription factors associated with bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bbx
|
ENSDARG00000012699 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bbx | dr11_v1_chr10_-_28525611_28525611 | -0.92 | 3.0e-08 | Click! |
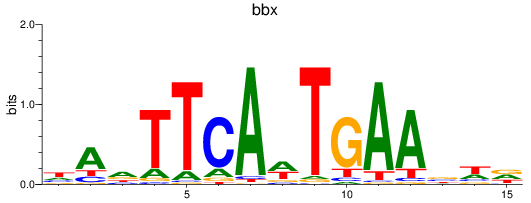
Activity profile of bbx motif
Sorted Z-values of bbx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_12031958 | 4.51 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr21_+_21263988 | 3.63 |
ENSDART00000089651
ENSDART00000108978 |
ccdc61
|
coiled-coil domain containing 61 |
| chr7_+_72003301 | 3.54 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr1_+_29858032 | 3.31 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr12_-_18568834 | 3.16 |
ENSDART00000039693
|
pgp
|
phosphoglycolate phosphatase |
| chr8_+_28593707 | 3.03 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr23_+_19670085 | 2.88 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr20_-_25626693 | 2.83 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr20_-_25626198 | 2.62 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr17_+_24718272 | 2.43 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr14_-_4177311 | 2.41 |
ENSDART00000128129
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr20_-_25626428 | 2.40 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr25_+_16646113 | 2.33 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr25_-_16076257 | 2.23 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr10_-_29892486 | 2.17 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr8_-_14091886 | 2.11 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr9_-_30555725 | 2.07 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr5_+_69733096 | 2.05 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr9_-_15424639 | 2.03 |
ENSDART00000124346
|
fn1a
|
fibronectin 1a |
| chr15_-_4596623 | 2.00 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr7_+_24729558 | 1.98 |
ENSDART00000111542
ENSDART00000170100 |
shroom4
|
shroom family member 4 |
| chr24_-_9002038 | 1.86 |
ENSDART00000066783
ENSDART00000150185 |
mppe1
|
metallophosphoesterase 1 |
| chr3_+_13862753 | 1.84 |
ENSDART00000168315
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr2_-_32237916 | 1.74 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr17_+_8799451 | 1.73 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr11_-_7410537 | 1.71 |
ENSDART00000009859
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr8_+_52701811 | 1.69 |
ENSDART00000179692
|
bcl2l16
|
BCL2 like 16 |
| chr17_+_8799661 | 1.68 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr8_-_40205712 | 1.67 |
ENSDART00000158927
|
anapc5
|
anaphase promoting complex subunit 5 |
| chr25_+_16880990 | 1.66 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr15_+_16886196 | 1.63 |
ENSDART00000139296
ENSDART00000049196 |
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr24_+_41989108 | 1.62 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr7_+_50395856 | 1.57 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr6_+_36839509 | 1.53 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr12_-_17686404 | 1.52 |
ENSDART00000079065
|
ccz1
|
CCZ1 homolog, vacuolar protein trafficking and biogenesis associated |
| chr15_-_35212462 | 1.51 |
ENSDART00000043960
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr11_+_2416064 | 1.51 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr16_-_27566552 | 1.47 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr13_-_24669258 | 1.42 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr7_+_42460302 | 1.42 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr3_+_54761569 | 1.40 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr6_+_30430591 | 1.38 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr12_-_33770299 | 1.37 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr11_-_12104917 | 1.36 |
ENSDART00000136108
|
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr6_+_37301341 | 1.33 |
ENSDART00000104180
|
zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr18_+_6857071 | 1.33 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr9_+_24106469 | 1.32 |
ENSDART00000039295
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr24_-_20926812 | 1.31 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr19_+_10860485 | 1.28 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
| chr23_+_35504824 | 1.25 |
ENSDART00000082647
ENSDART00000159218 |
acp1
|
acid phosphatase 1 |
| chr19_-_438337 | 1.25 |
ENSDART00000127985
|
MTERF1
|
mitochondrial transcription termination factor 1 |
| chr7_+_36898850 | 1.24 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr14_+_36223097 | 1.21 |
ENSDART00000186872
|
pitx2
|
paired-like homeodomain 2 |
| chr1_+_35956435 | 1.20 |
ENSDART00000085021
ENSDART00000148505 |
mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr6_+_6780873 | 1.20 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr15_+_23528310 | 1.19 |
ENSDART00000152523
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr8_+_31016180 | 1.18 |
ENSDART00000130870
ENSDART00000143604 |
odf2b
|
outer dense fiber of sperm tails 2b |
| chr16_-_47427016 | 1.18 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr16_+_32014552 | 1.15 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr13_+_18507592 | 1.15 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr21_-_25416391 | 1.12 |
ENSDART00000114081
|
sms
|
spermine synthase |
| chr8_+_21114338 | 1.12 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| chr15_+_36310442 | 1.12 |
ENSDART00000154678
|
gmnc
|
geminin coiled-coil domain containing |
| chr1_-_7673376 | 1.11 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr1_+_12767318 | 1.10 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr4_+_5215950 | 1.09 |
ENSDART00000140613
|
galnt8b.2
|
polypeptide N-acetylgalactosaminyltransferase 8b, tandem duplicate 2 |
| chr23_-_30781875 | 1.09 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr20_-_20248408 | 1.07 |
ENSDART00000183234
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr6_-_43922813 | 1.06 |
ENSDART00000123341
|
prok2
|
prokineticin 2 |
| chr11_-_23322182 | 1.06 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr2_-_22659450 | 1.04 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr17_+_43867889 | 1.03 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr1_+_55643198 | 1.01 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr5_+_36650096 | 1.00 |
ENSDART00000111414
|
alkbh6
|
alkB homolog 6 |
| chr12_+_27156943 | 1.00 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr21_+_38732945 | 1.00 |
ENSDART00000076157
|
rab24
|
RAB24, member RAS oncogene family |
| chr23_+_17512037 | 0.99 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr22_+_9939901 | 0.98 |
ENSDART00000169777
ENSDART00000081420 |
zgc:171686
|
zgc:171686 |
| chr1_-_28831848 | 0.97 |
ENSDART00000148536
|
GK3P
|
zgc:172295 |
| chr20_+_25626479 | 0.97 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr21_+_21709204 | 0.96 |
ENSDART00000137478
|
or125-1
|
odorant receptor, family E, subfamily 125, member 1 |
| chr11_-_27501027 | 0.95 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr8_+_23784471 | 0.94 |
ENSDART00000189457
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr13_-_7233811 | 0.94 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr14_+_36246726 | 0.90 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr14_-_29826659 | 0.89 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr23_-_30781510 | 0.85 |
ENSDART00000183364
|
myt1a
|
myelin transcription factor 1a |
| chr12_-_9700605 | 0.83 |
ENSDART00000161063
|
heatr1
|
HEAT repeat containing 1 |
| chr6_+_14980761 | 0.82 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr10_-_43771447 | 0.81 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr18_-_17724295 | 0.80 |
ENSDART00000121553
|
fam192a
|
family with sequence similarity 192, member A |
| chr15_-_31419805 | 0.80 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr18_-_26510545 | 0.80 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr3_-_34816893 | 0.78 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr25_+_23591990 | 0.78 |
ENSDART00000151938
ENSDART00000089199 |
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr16_-_24195252 | 0.78 |
ENSDART00000136205
ENSDART00000048599 ENSDART00000161547 |
rps19
|
ribosomal protein S19 |
| chr1_-_45039726 | 0.75 |
ENSDART00000186188
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr4_+_16323970 | 0.75 |
ENSDART00000190651
|
BX322608.1
|
|
| chr1_+_2431956 | 0.74 |
ENSDART00000183832
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr11_-_18323059 | 0.71 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr1_+_19535144 | 0.71 |
ENSDART00000103089
|
si:dkey-245p14.4
|
si:dkey-245p14.4 |
| chr4_+_60296027 | 0.69 |
ENSDART00000172479
|
si:dkey-248e17.5
|
si:dkey-248e17.5 |
| chr18_+_26899316 | 0.69 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr9_+_48755638 | 0.68 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr12_-_31484677 | 0.67 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr5_+_37744625 | 0.65 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr21_+_29077509 | 0.62 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr17_+_23549117 | 0.61 |
ENSDART00000063953
|
zgc:65997
|
zgc:65997 |
| chr4_+_47636303 | 0.60 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr16_-_30826712 | 0.59 |
ENSDART00000122474
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr8_-_22157301 | 0.57 |
ENSDART00000158383
|
nphp4
|
nephronophthisis 4 |
| chr6_-_40921412 | 0.57 |
ENSDART00000076097
ENSDART00000188364 |
sfi1
|
SFI1 centrin binding protein |
| chr1_-_46244523 | 0.56 |
ENSDART00000143908
|
si:ch211-138g9.3
|
si:ch211-138g9.3 |
| chr19_+_33093395 | 0.55 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr1_+_23784905 | 0.55 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr21_-_22647695 | 0.54 |
ENSDART00000187553
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr25_-_12730260 | 0.53 |
ENSDART00000171834
|
klhdc4
|
kelch domain containing 4 |
| chr10_-_34107919 | 0.51 |
ENSDART00000167428
ENSDART00000145279 |
pimr144
pimr146
|
Pim proto-oncogene, serine/threonine kinase, related 144 Pim proto-oncogene, serine/threonine kinase, related 146 |
| chr8_+_23034718 | 0.51 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr3_-_37759969 | 0.50 |
ENSDART00000151105
ENSDART00000151208 |
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr2_-_10188598 | 0.49 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr16_-_13622794 | 0.48 |
ENSDART00000146953
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr7_-_50395395 | 0.48 |
ENSDART00000065868
|
vps33b
|
vacuolar protein sorting 33B |
| chr6_-_35052388 | 0.46 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr25_+_22107643 | 0.46 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr4_-_5595237 | 0.44 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr1_-_9104631 | 0.44 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr15_-_5222924 | 0.44 |
ENSDART00000128924
|
or128-4
|
odorant receptor, family E, subfamily 128, member 4 |
| chr4_+_47768097 | 0.43 |
ENSDART00000192738
|
BX324142.2
|
|
| chr19_+_33093577 | 0.43 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr17_-_8656155 | 0.41 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr7_+_10701770 | 0.41 |
ENSDART00000167323
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr16_-_45235947 | 0.41 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr16_+_16977786 | 0.41 |
ENSDART00000043173
ENSDART00000132150 |
rpl18
|
ribosomal protein L18 |
| chr14_+_21005372 | 0.40 |
ENSDART00000170638
|
pimr195
|
Pim proto-oncogene, serine/threonine kinase, related 195 |
| chr13_+_6086730 | 0.39 |
ENSDART00000049328
|
fam120b
|
family with sequence similarity 120B |
| chr8_+_20438884 | 0.39 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr15_-_1022436 | 0.38 |
ENSDART00000156003
|
znf1010
|
zinc finger protein 1010 |
| chr14_-_30808174 | 0.37 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr2_+_19633493 | 0.36 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr16_-_12291045 | 0.35 |
ENSDART00000185447
|
CR388102.1
|
|
| chr3_-_12026741 | 0.34 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr7_-_50395059 | 0.34 |
ENSDART00000191150
|
vps33b
|
vacuolar protein sorting 33B |
| chr8_+_20951590 | 0.33 |
ENSDART00000138728
|
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr16_+_16978424 | 0.32 |
ENSDART00000143128
|
rpl18
|
ribosomal protein L18 |
| chr18_+_17493859 | 0.27 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr10_-_42751641 | 0.25 |
ENSDART00000182734
ENSDART00000113926 |
zgc:100918
|
zgc:100918 |
| chr11_+_10548171 | 0.25 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr22_+_35930526 | 0.24 |
ENSDART00000169242
|
LO016987.1
|
|
| chr8_+_3431671 | 0.21 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr23_+_2703044 | 0.20 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr23_-_18381361 | 0.18 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr22_+_19230434 | 0.18 |
ENSDART00000132929
|
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr17_-_24439672 | 0.16 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr13_-_50565338 | 0.15 |
ENSDART00000062684
|
blnk
|
B cell linker |
| chr1_+_55583116 | 0.15 |
ENSDART00000152163
|
adgre19
|
adhesion G protein-coupled receptor E19 |
| chr13_-_23095006 | 0.14 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr6_-_54816567 | 0.14 |
ENSDART00000150079
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr25_-_16070806 | 0.13 |
ENSDART00000162645
|
CR759887.3
|
|
| chr12_+_23812530 | 0.12 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr6_-_19042294 | 0.09 |
ENSDART00000159461
|
si:rp71-81e14.2
|
si:rp71-81e14.2 |
| chr19_+_43341424 | 0.09 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr1_+_55535827 | 0.07 |
ENSDART00000152784
|
adgre16
|
adhesion G protein-coupled receptor E16 |
| chr7_-_1163829 | 0.07 |
ENSDART00000181937
|
CT030017.1
|
|
| chr20_+_3934516 | 0.07 |
ENSDART00000165732
|
clec11a
|
C-type lectin domain containing 11A |
| chr15_+_44201056 | 0.06 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr4_+_76442674 | 0.06 |
ENSDART00000164825
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_+_32553714 | 0.04 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr2_-_41518340 | 0.02 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr9_+_19623363 | 0.01 |
ENSDART00000142471
ENSDART00000147662 ENSDART00000136053 |
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr1_+_55563532 | 0.00 |
ENSDART00000152549
|
adgre15
|
adhesion G protein-coupled receptor E15 |
| chr13_+_23095228 | 0.00 |
ENSDART00000189068
ENSDART00000188624 |
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bbx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 1.6 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.9 | 8.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.8 | 3.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.5 | 3.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.4 | 2.1 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.3 | 2.2 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.3 | 2.0 | GO:0045198 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) acute-phase response(GO:0006953) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 1.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 4.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 2.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.3 | 1.4 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 1.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 1.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.7 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 3.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 1.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.8 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 3.6 | GO:0098534 | centriole assembly(GO:0098534) |
| 0.1 | 1.4 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 0.9 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 1.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.1 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.8 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.8 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 4.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 1.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.6 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 1.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 2.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.7 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.1 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 1.1 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.4 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 2.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.6 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 1.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.0 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 1.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.0 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 2.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.7 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.0 | 2.5 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.7 | GO:0048840 | otolith development(GO:0048840) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 3.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 3.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 1.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.3 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.2 | 2.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 3.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.2 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.5 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.7 | 2.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 1.3 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.3 | 2.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.3 | 2.0 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.3 | 1.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 1.7 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.7 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.5 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.3 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 1.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.2 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 1.1 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.9 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 4.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 4.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.8 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |