Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
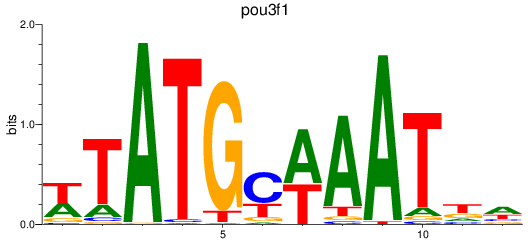
Results for pou3f1
Z-value: 2.08
Transcription factors associated with pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f1
|
ENSDARG00000009823 | POU class 3 homeobox 1 |
|
pou3f1
|
ENSDARG00000112302 | POU class 3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f1 | dr11_v1_chr16_+_33593116_33593116 | 0.94 | 1.6e-04 | Click! |
Activity profile of pou3f1 motif
Sorted Z-values of pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_31372639 | 6.48 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr6_+_4539953 | 4.97 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr3_-_32817274 | 4.78 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr21_+_30563115 | 3.98 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr21_+_26390549 | 3.74 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr9_+_54178475 | 3.34 |
ENSDART00000104475
|
tmsb4x
|
thymosin, beta 4 x |
| chr23_+_21473103 | 3.22 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr2_+_55982940 | 3.20 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr4_-_16406737 | 3.02 |
ENSDART00000013085
|
dcn
|
decorin |
| chr8_+_13503377 | 2.98 |
ENSDART00000034740
ENSDART00000167187 |
fut9d
|
fucosyltransferase 9d |
| chr13_+_33117528 | 2.96 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr7_-_24699985 | 2.94 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr1_+_54013457 | 2.93 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr9_-_33877476 | 2.89 |
ENSDART00000150035
ENSDART00000088441 ENSDART00000183210 |
si:ch73-147f11.1
|
si:ch73-147f11.1 |
| chr17_+_33719415 | 2.83 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr9_-_18877597 | 2.77 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr1_+_46194333 | 2.74 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr3_-_5964557 | 2.74 |
ENSDART00000184738
|
BX284638.1
|
|
| chr19_+_3653976 | 2.73 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr3_+_23743139 | 2.71 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr7_+_40228422 | 2.66 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr4_-_9592402 | 2.65 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr5_-_42272517 | 2.64 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr22_+_5106751 | 2.57 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr18_+_38749547 | 2.56 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr20_+_40150612 | 2.56 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr25_+_34576067 | 2.48 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr19_+_19759577 | 2.39 |
ENSDART00000169480
|
hoxa5a
|
homeobox A5a |
| chr23_+_36771593 | 2.35 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr10_+_32104305 | 2.32 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr13_+_29778610 | 2.26 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr23_-_3409140 | 2.24 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr5_+_58550291 | 2.22 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr22_+_15624371 | 2.18 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr19_+_19775757 | 2.17 |
ENSDART00000164677
|
hoxa3a
|
homeobox A3a |
| chr13_+_30951155 | 2.05 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr16_-_12173554 | 2.04 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr19_-_2318391 | 2.03 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr18_+_44649804 | 1.96 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr13_-_6081803 | 1.96 |
ENSDART00000099224
|
dld
|
deltaD |
| chr21_+_41743493 | 1.94 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr5_-_25406807 | 1.93 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr19_+_19786117 | 1.92 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr25_-_15045338 | 1.87 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr23_+_28731379 | 1.86 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr5_-_24000211 | 1.85 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr6_-_13187168 | 1.83 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr3_+_23697997 | 1.83 |
ENSDART00000184299
ENSDART00000078466 |
hoxb3a
|
homeobox B3a |
| chr19_+_3001473 | 1.83 |
ENSDART00000145363
|
scxa
|
scleraxis bHLH transcription factor a |
| chr17_-_32865788 | 1.80 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr22_+_20195280 | 1.76 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr23_-_20258490 | 1.74 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr9_-_21507083 | 1.73 |
ENSDART00000137922
|
aff3
|
AF4/FMR2 family, member 3 |
| chr3_+_23742868 | 1.69 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr13_-_33822550 | 1.68 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr7_-_39537292 | 1.67 |
ENSDART00000173965
|
otog
|
otogelin |
| chr1_+_7679328 | 1.66 |
ENSDART00000163488
ENSDART00000190070 |
en1b
|
engrailed homeobox 1b |
| chr18_+_22606259 | 1.66 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr20_+_48782068 | 1.65 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr7_+_25920792 | 1.65 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr14_+_36889893 | 1.64 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr14_-_26704829 | 1.62 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr10_+_22003750 | 1.61 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr6_+_41096058 | 1.61 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr25_-_13188678 | 1.58 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr18_+_29402623 | 1.58 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr18_-_36135799 | 1.56 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr23_+_26733232 | 1.55 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr23_+_28582865 | 1.54 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr6_-_13188667 | 1.53 |
ENSDART00000191654
|
adam23a
|
ADAM metallopeptidase domain 23a |
| chr4_+_31259 | 1.53 |
ENSDART00000166826
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr14_-_40389699 | 1.52 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr23_-_3408777 | 1.50 |
ENSDART00000193245
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr11_+_25257022 | 1.50 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr20_-_39735952 | 1.46 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr19_+_2726819 | 1.42 |
ENSDART00000187122
ENSDART00000112414 |
rapgef5a
|
Rap guanine nucleotide exchange factor (GEF) 5a |
| chr11_-_44876005 | 1.39 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr21_-_35853245 | 1.38 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr8_+_28593707 | 1.38 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr20_-_39188476 | 1.35 |
ENSDART00000152858
|
rcan2
|
regulator of calcineurin 2 |
| chr13_+_28705143 | 1.30 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr7_-_4036184 | 1.30 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr5_+_29820266 | 1.29 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr18_+_29403017 | 1.27 |
ENSDART00000176966
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr16_+_43152727 | 1.26 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr16_-_12173399 | 1.26 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr17_-_42213822 | 1.25 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr19_-_30562648 | 1.25 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr10_+_36458563 | 1.25 |
ENSDART00000077008
|
alox5ap
|
arachidonate 5-lipoxygenase-activating protein |
| chr19_+_14573998 | 1.24 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr8_-_12403077 | 1.24 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr2_+_52847049 | 1.23 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr17_-_37052622 | 1.23 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr1_-_17587552 | 1.22 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr5_+_22970617 | 1.21 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr19_-_28367413 | 1.20 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr4_+_12013043 | 1.20 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr13_-_7031033 | 1.15 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr9_-_1984604 | 1.15 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr21_+_5888641 | 1.11 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr10_+_33895315 | 1.11 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr14_-_48939560 | 1.10 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr11_+_25634041 | 1.10 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr7_-_30367650 | 1.07 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr24_-_8730913 | 1.06 |
ENSDART00000187228
ENSDART00000082349 ENSDART00000186660 |
tfap2a
|
transcription factor AP-2 alpha |
| chr15_-_18361475 | 1.05 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr18_-_42830563 | 1.05 |
ENSDART00000191488
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr9_-_9980704 | 1.04 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr12_-_38548299 | 1.04 |
ENSDART00000153374
|
si:dkey-1f1.3
|
si:dkey-1f1.3 |
| chr5_-_46896541 | 1.04 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr6_-_16406210 | 1.03 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr12_+_23424108 | 1.03 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr4_+_12013642 | 1.03 |
ENSDART00000067281
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr25_-_25736958 | 1.02 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr13_+_29773153 | 1.01 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr21_-_5393125 | 1.00 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr21_+_38817785 | 1.00 |
ENSDART00000177616
ENSDART00000149085 |
hnf1bb
|
HNF1 homeobox Bb |
| chr22_+_17784414 | 0.99 |
ENSDART00000188189
ENSDART00000145260 |
tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr3_-_32337653 | 0.98 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr25_-_16057539 | 0.96 |
ENSDART00000090388
|
cyb5r2
|
cytochrome b5 reductase 2 |
| chr6_+_16406723 | 0.92 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr3_+_47322494 | 0.91 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr15_-_16012963 | 0.90 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr1_+_26411496 | 0.88 |
ENSDART00000112263
|
arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr7_+_61050329 | 0.87 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr10_+_31248036 | 0.87 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr8_+_28469054 | 0.86 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr10_-_27196093 | 0.85 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr20_+_35998073 | 0.84 |
ENSDART00000140457
|
opn5
|
opsin 5 |
| chr6_+_23935045 | 0.84 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr1_+_1599979 | 0.83 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr14_-_28001986 | 0.83 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr16_-_31756859 | 0.82 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_+_69470442 | 0.81 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr23_+_44307996 | 0.81 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr22_-_32392252 | 0.81 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr4_-_11580948 | 0.80 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr22_-_18491813 | 0.79 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr24_+_25467465 | 0.79 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr7_+_60396957 | 0.79 |
ENSDART00000121545
|
brms1
|
breast cancer metastasis suppressor 1 |
| chr10_+_453619 | 0.78 |
ENSDART00000135598
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr15_-_14212777 | 0.78 |
ENSDART00000165572
|
CR925813.1
|
|
| chr2_-_34555945 | 0.78 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr17_+_15882533 | 0.77 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr23_+_21380079 | 0.74 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr1_+_5275811 | 0.74 |
ENSDART00000189676
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr15_+_39096736 | 0.73 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr5_-_54497319 | 0.72 |
ENSDART00000160492
|
alad
|
aminolevulinate dehydratase |
| chr12_-_34887943 | 0.72 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr1_-_12109216 | 0.71 |
ENSDART00000079930
|
mttp
|
microsomal triglyceride transfer protein |
| chr25_-_6557854 | 0.70 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr15_+_45640906 | 0.68 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr22_-_11729350 | 0.68 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr23_+_11347313 | 0.67 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr2_+_30147504 | 0.66 |
ENSDART00000190947
|
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr12_+_2677303 | 0.66 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr14_-_34700633 | 0.65 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr18_-_49286381 | 0.65 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr8_-_15398760 | 0.64 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr2_+_33382648 | 0.64 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_-_24916889 | 0.64 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr3_-_31783737 | 0.63 |
ENSDART00000090809
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr11_-_35575791 | 0.63 |
ENSDART00000031441
ENSDART00000188513 ENSDART00000183609 |
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr25_+_36674715 | 0.62 |
ENSDART00000111861
|
mafb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
| chr19_-_6239248 | 0.62 |
ENSDART00000014127
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr5_-_24238733 | 0.62 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr25_+_19666621 | 0.60 |
ENSDART00000112746
|
zgc:171459
|
zgc:171459 |
| chr16_+_25535993 | 0.60 |
ENSDART00000077436
|
mylipb
|
myosin regulatory light chain interacting protein b |
| chr1_-_20593778 | 0.59 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr2_-_24317240 | 0.58 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr17_-_30473374 | 0.58 |
ENSDART00000155021
|
si:ch211-175f11.5
|
si:ch211-175f11.5 |
| chr2_+_32835679 | 0.57 |
ENSDART00000132767
|
pxdc1a
|
PX domain containing 1a |
| chr5_-_16996482 | 0.57 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr22_+_5135884 | 0.56 |
ENSDART00000141276
|
mydgf
|
myeloid-derived growth factor |
| chr16_+_40560622 | 0.56 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr1_-_1402303 | 0.54 |
ENSDART00000130697
|
runx1
|
runt-related transcription factor 1 |
| chr4_+_20255160 | 0.54 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr7_-_38633867 | 0.54 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr18_+_7286788 | 0.53 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr20_-_25626693 | 0.53 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr16_+_45746549 | 0.53 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr7_-_40738774 | 0.52 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr16_-_21620947 | 0.52 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr23_-_1017605 | 0.51 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr5_-_63302944 | 0.51 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr10_-_24689280 | 0.50 |
ENSDART00000191476
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr10_-_5821584 | 0.50 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr1_+_31725154 | 0.49 |
ENSDART00000112333
ENSDART00000189801 |
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr24_+_41931585 | 0.48 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr14_-_12391506 | 0.46 |
ENSDART00000080864
|
magt1
|
magnesium transporter 1 |
| chr3_+_33345348 | 0.46 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr2_-_56131312 | 0.45 |
ENSDART00000097755
|
jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr23_+_32011768 | 0.45 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr1_+_27825980 | 0.44 |
ENSDART00000160524
|
pspc1
|
paraspeckle component 1 |
| chr7_-_23873173 | 0.44 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr5_+_38837429 | 0.44 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr5_+_30624183 | 0.43 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr21_-_27185915 | 0.42 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr3_-_50998577 | 0.41 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr24_-_32452355 | 0.41 |
ENSDART00000146511
|
ccdc129
|
coiled-coil domain containing 129 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.9 | 3.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.9 | 3.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.9 | 4.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.7 | 2.9 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.6 | 1.8 | GO:0035992 | tendon formation(GO:0035992) |
| 0.6 | 1.7 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.5 | 5.2 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.5 | 3.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.5 | 2.9 | GO:0055016 | hypochord development(GO:0055016) |
| 0.4 | 1.3 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 | 1.7 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.4 | 2.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.4 | 3.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 5.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 1.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.3 | 2.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 1.8 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.3 | 0.9 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.3 | 0.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.3 | 1.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 1.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 1.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 1.2 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 2.6 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.8 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.2 | 5.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 2.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 1.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 0.6 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 0.6 | GO:0060306 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.2 | 1.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 1.7 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 0.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.5 | GO:0010755 | regulation of collagen metabolic process(GO:0010712) regulation of plasminogen activation(GO:0010755) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 3.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 2.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.5 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 1.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 2.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.5 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.1 | 1.0 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.8 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.3 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.1 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) |
| 0.1 | 0.7 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.1 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.8 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.7 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.5 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.4 | GO:0031670 | cellular response to nutrient(GO:0031670) vitamin D3 metabolic process(GO:0070640) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
| 0.1 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 1.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 2.5 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 1.0 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.4 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 1.7 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 1.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.6 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 1.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.8 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 2.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 6.9 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.4 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.6 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.1 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.0 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.0 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.1 | GO:1901021 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of calcium ion transmembrane transport(GO:1904427) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.8 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.4 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.2 | 1.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 4.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 2.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.7 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 3.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 3.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.9 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.6 | 1.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.5 | 1.6 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.5 | 3.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.4 | 1.7 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.4 | 1.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 1.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.4 | 7.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 7.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 5.8 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.3 | 1.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 4.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 0.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.3 | 1.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 2.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 4.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.7 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.2 | 1.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.5 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.4 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 0.8 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 3.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.9 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 15.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 27.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 5.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 3.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 0.8 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 2.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |