Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
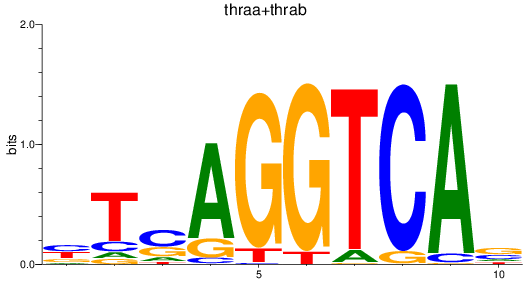
Results for thraa+thrab
Z-value: 1.74
Transcription factors associated with thraa+thrab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
thraa
|
ENSDARG00000000151 | thyroid hormone receptor alpha a |
|
thrab
|
ENSDARG00000052654 | thyroid hormone receptor alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| thraa | dr11_v1_chr3_-_34724879_34724879 | -0.71 | 3.4e-02 | Click! |
| thrab | dr11_v1_chr12_-_22039350_22039350 | -0.29 | 4.6e-01 | Click! |
Activity profile of thraa+thrab motif
Sorted Z-values of thraa+thrab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_39185761 | 5.76 |
ENSDART00000075992
ENSDART00000140644 |
cryba1b
|
crystallin, beta A1b |
| chr9_-_22129788 | 3.66 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr6_-_13783604 | 3.66 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr10_-_44027391 | 3.57 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr21_+_39185461 | 3.55 |
ENSDART00000178419
|
cryba1b
|
crystallin, beta A1b |
| chr9_-_22135576 | 3.33 |
ENSDART00000101902
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr9_-_22272181 | 3.29 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr9_-_22245572 | 3.12 |
ENSDART00000114943
|
crygm2d4
|
crystallin, gamma M2d4 |
| chr20_-_40717900 | 3.01 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr12_-_30841679 | 2.87 |
ENSDART00000105594
|
crygmx
|
crystallin, gamma MX |
| chr9_-_22299412 | 2.77 |
ENSDART00000139101
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr9_-_22147567 | 2.74 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr1_+_29096881 | 2.63 |
ENSDART00000075539
|
cryaa
|
crystallin, alpha A |
| chr9_-_22205682 | 2.32 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr9_-_22240052 | 2.27 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr7_+_26716321 | 2.17 |
ENSDART00000189750
|
cd82a
|
CD82 molecule a |
| chr2_+_42191592 | 2.07 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr3_-_38915998 | 2.04 |
ENSDART00000141886
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr1_+_16127825 | 1.96 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr9_-_22023061 | 1.88 |
ENSDART00000101952
|
crygm2c
|
crystallin, gamma M2c |
| chr19_-_47456787 | 1.78 |
ENSDART00000168792
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr13_-_16257848 | 1.74 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr12_-_3133483 | 1.73 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr3_+_33300522 | 1.73 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr7_+_20017211 | 1.70 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr24_-_39610585 | 1.63 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr3_-_18575868 | 1.59 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr19_+_3653976 | 1.57 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr13_-_31435137 | 1.56 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr10_+_6318227 | 1.51 |
ENSDART00000170872
ENSDART00000162428 ENSDART00000158994 |
tpm2
|
tropomyosin 2 (beta) |
| chr17_+_23300827 | 1.49 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr7_-_30082931 | 1.49 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr19_+_1184878 | 1.47 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr4_-_16853464 | 1.46 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr4_-_1360495 | 1.45 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr16_+_33655890 | 1.44 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr11_-_31172276 | 1.44 |
ENSDART00000171520
|
si:dkey-238i5.3
|
si:dkey-238i5.3 |
| chr21_-_41870029 | 1.40 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr13_+_22249636 | 1.37 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr11_+_23265157 | 1.35 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr16_+_5774977 | 1.35 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr7_-_41627956 | 1.34 |
ENSDART00000083929
|
plxdc2
|
plexin domain containing 2 |
| chr8_-_16697615 | 1.33 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr11_-_44543082 | 1.32 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr1_-_59176949 | 1.26 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr15_+_28096152 | 1.25 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr13_-_30645965 | 1.22 |
ENSDART00000109307
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr4_+_42604252 | 1.22 |
ENSDART00000184850
|
CR925768.1
|
|
| chr20_+_30490682 | 1.22 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr19_+_10341738 | 1.22 |
ENSDART00000128721
|
rcvrn3
|
recoverin 3 |
| chr10_+_22782522 | 1.21 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr8_-_2434282 | 1.21 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr7_+_31871830 | 1.20 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr13_-_36703164 | 1.19 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr19_+_30662529 | 1.19 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr11_+_45421761 | 1.19 |
ENSDART00000167347
|
hrasls
|
HRAS-like suppressor |
| chr13_-_29424454 | 1.19 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr25_-_5740334 | 1.18 |
ENSDART00000169622
ENSDART00000168720 |
LO017739.1
|
|
| chr13_-_39947335 | 1.18 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr6_+_41463786 | 1.18 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr20_+_54738210 | 1.17 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr7_-_17028015 | 1.16 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr23_-_11870962 | 1.16 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr2_+_33368414 | 1.14 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr20_-_26001288 | 1.14 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr18_-_48492951 | 1.14 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr18_-_15373620 | 1.11 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr23_-_16843649 | 1.10 |
ENSDART00000129971
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr7_+_29133321 | 1.10 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr3_+_23737795 | 1.10 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr18_+_36037223 | 1.06 |
ENSDART00000144410
|
tmem91
|
transmembrane protein 91 |
| chr14_+_36231126 | 1.05 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr4_+_30776883 | 1.03 |
ENSDART00000169519
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr6_+_1787160 | 1.02 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr23_-_4855122 | 1.01 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr25_-_29134654 | 1.01 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr4_-_16412084 | 1.00 |
ENSDART00000188460
|
dcn
|
decorin |
| chr4_+_42450386 | 0.99 |
ENSDART00000168211
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr3_-_29977495 | 0.99 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr16_+_50289916 | 0.98 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr9_-_22892838 | 0.96 |
ENSDART00000143888
|
neb
|
nebulin |
| chr17_-_27273296 | 0.96 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr6_-_60031693 | 0.96 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr11_+_45422206 | 0.89 |
ENSDART00000182548
|
hrasls
|
HRAS-like suppressor |
| chr21_+_25688388 | 0.89 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr8_+_25902170 | 0.88 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr15_+_24644016 | 0.88 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr4_+_38344 | 0.87 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr11_-_30138299 | 0.87 |
ENSDART00000172106
|
scml2
|
Scm polycomb group protein like 2 |
| chr5_-_51747278 | 0.86 |
ENSDART00000192232
|
lhfpl2b
|
LHFPL tetraspan subfamily member 2b |
| chr16_+_22618620 | 0.85 |
ENSDART00000185728
ENSDART00000041625 |
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr9_+_907459 | 0.83 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr9_+_54644626 | 0.83 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr11_+_25477643 | 0.81 |
ENSDART00000065941
|
opn1lw1
|
opsin 1 (cone pigments), long-wave-sensitive, 1 |
| chr18_-_48517040 | 0.80 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr19_-_103289 | 0.79 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr19_-_24443394 | 0.79 |
ENSDART00000090825
|
thbs3b
|
thrombospondin 3b |
| chr7_+_36035432 | 0.79 |
ENSDART00000179004
|
irx3a
|
iroquois homeobox 3a |
| chr14_-_7128980 | 0.77 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr11_-_11471857 | 0.77 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr10_-_44341288 | 0.75 |
ENSDART00000166131
|
zswim6
|
zinc finger, SWIM-type containing 6 |
| chr18_-_15932704 | 0.75 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr5_-_33286820 | 0.75 |
ENSDART00000184426
|
rpl7a
|
ribosomal protein L7a |
| chr18_-_48508585 | 0.74 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr14_-_42997145 | 0.74 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr19_+_4912817 | 0.74 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_-_26924903 | 0.73 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr4_+_75314247 | 0.71 |
ENSDART00000162365
|
CABZ01041913.1
|
|
| chr10_-_39153959 | 0.69 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr10_+_8629275 | 0.69 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr10_-_39154594 | 0.69 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr3_-_36612877 | 0.69 |
ENSDART00000167164
|
si:dkeyp-72e1.7
|
si:dkeyp-72e1.7 |
| chr15_-_35406564 | 0.68 |
ENSDART00000086051
|
mecom
|
MDS1 and EVI1 complex locus |
| chr11_-_1400507 | 0.68 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr5_-_29643381 | 0.68 |
ENSDART00000034849
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr21_-_45073764 | 0.68 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr19_-_24443867 | 0.67 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr1_+_34696503 | 0.66 |
ENSDART00000186106
|
CR339054.2
|
|
| chr18_+_8340886 | 0.64 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr16_-_46605160 | 0.63 |
ENSDART00000189070
ENSDART00000181194 |
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr2_-_28102264 | 0.63 |
ENSDART00000013638
|
cdh6
|
cadherin 6 |
| chr25_+_10830269 | 0.62 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr3_+_12484008 | 0.62 |
ENSDART00000182229
|
vasnb
|
vasorin b |
| chr8_+_28259347 | 0.61 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr3_+_58379450 | 0.61 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr23_+_30048849 | 0.61 |
ENSDART00000126027
|
uts2a
|
urotensin 2, alpha |
| chr5_+_15350954 | 0.61 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr3_-_45777226 | 0.59 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr17_+_24613255 | 0.58 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr8_-_25980694 | 0.58 |
ENSDART00000135456
|
si:dkey-72l14.3
|
si:dkey-72l14.3 |
| chr1_-_45347393 | 0.58 |
ENSDART00000173024
|
si:ch211-243a20.4
|
si:ch211-243a20.4 |
| chr12_+_34896956 | 0.57 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr5_-_30535327 | 0.57 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr15_-_28095532 | 0.57 |
ENSDART00000191490
|
cryba1a
|
crystallin, beta A1a |
| chr23_+_42810055 | 0.56 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr4_-_77432218 | 0.56 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr5_+_27440325 | 0.56 |
ENSDART00000185815
ENSDART00000144013 |
loxl2b
|
lysyl oxidase-like 2b |
| chr8_-_47818282 | 0.56 |
ENSDART00000137149
|
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr4_+_17280868 | 0.56 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr8_-_16712111 | 0.55 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr1_+_23783349 | 0.55 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr4_+_15899668 | 0.55 |
ENSDART00000101592
|
col7a1l
|
collagen type VII alpha 1-like |
| chr2_-_44199722 | 0.54 |
ENSDART00000140633
ENSDART00000145728 |
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr7_-_72435159 | 0.54 |
ENSDART00000173060
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr25_-_15214161 | 0.54 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr17_+_389218 | 0.54 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr23_-_3703569 | 0.53 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr2_+_6181383 | 0.53 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr20_+_45893173 | 0.52 |
ENSDART00000131169
|
bmp2b
|
bone morphogenetic protein 2b |
| chr10_+_35439952 | 0.52 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr13_+_51869025 | 0.51 |
ENSDART00000187066
|
LT631684.1
|
|
| chr16_+_53125918 | 0.51 |
ENSDART00000102170
|
CABZ01053976.1
|
|
| chr23_+_4373360 | 0.51 |
ENSDART00000144061
|
ptpdc1b
|
protein tyrosine phosphatase domain containing 1b |
| chr18_+_48943875 | 0.50 |
ENSDART00000076803
|
FO681288.1
|
|
| chr8_+_25352268 | 0.50 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr1_-_45177373 | 0.49 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr20_-_7992496 | 0.48 |
ENSDART00000145521
|
plpp3
|
phospholipid phosphatase 3 |
| chr25_-_36492779 | 0.48 |
ENSDART00000042271
|
irx3b
|
iroquois homeobox 3b |
| chr9_-_11550711 | 0.48 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr13_+_23677949 | 0.47 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr16_-_5721386 | 0.46 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr4_-_37479395 | 0.46 |
ENSDART00000168299
ENSDART00000157864 |
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr1_-_22512063 | 0.46 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr14_-_30390145 | 0.45 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr4_+_26496489 | 0.45 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr7_-_26308098 | 0.45 |
ENSDART00000146440
ENSDART00000146935 ENSDART00000164627 |
zgc:77439
|
zgc:77439 |
| chr16_+_5678071 | 0.45 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr9_+_17429170 | 0.44 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr1_-_30039331 | 0.44 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr7_-_37812176 | 0.43 |
ENSDART00000164485
|
adcy7
|
adenylate cyclase 7 |
| chr18_+_45542981 | 0.43 |
ENSDART00000140357
|
kifc3
|
kinesin family member C3 |
| chr20_+_5564042 | 0.43 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr5_-_23277385 | 0.43 |
ENSDART00000134982
|
plp1b
|
proteolipid protein 1b |
| chr16_-_44649053 | 0.43 |
ENSDART00000184807
|
CR925804.2
|
|
| chr5_+_23024033 | 0.43 |
ENSDART00000171199
|
chmp1b
|
chromatin modifying protein 1B |
| chr14_-_24332786 | 0.43 |
ENSDART00000173164
|
fam13b
|
family with sequence similarity 13, member B |
| chr18_-_26785861 | 0.42 |
ENSDART00000098361
|
nmba
|
neuromedin Ba |
| chr10_+_22775253 | 0.41 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr20_+_36623807 | 0.41 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr9_+_21281652 | 0.41 |
ENSDART00000113352
|
lats2
|
large tumor suppressor kinase 2 |
| chr2_-_4797512 | 0.41 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr5_-_58840971 | 0.40 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr11_+_6819050 | 0.40 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr6_+_29305190 | 0.40 |
ENSDART00000078647
|
si:ch211-201h21.5
|
si:ch211-201h21.5 |
| chr2_+_24786765 | 0.39 |
ENSDART00000141030
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr14_-_34771371 | 0.39 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr8_-_7391721 | 0.39 |
ENSDART00000149836
|
lhfpl4b
|
LHFPL tetraspan subfamily member 4b |
| chr5_-_23277939 | 0.38 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr24_+_26036395 | 0.38 |
ENSDART00000143485
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr20_+_19793620 | 0.38 |
ENSDART00000136317
|
chrna2b
|
cholinergic receptor, nicotinic, alpha 2b (neuronal) |
| chr1_-_38813679 | 0.38 |
ENSDART00000148917
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr20_-_9755546 | 0.37 |
ENSDART00000152498
ENSDART00000152239 |
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr24_-_7995748 | 0.37 |
ENSDART00000158566
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr4_-_25271455 | 0.37 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr12_+_7491690 | 0.37 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr24_-_7995960 | 0.36 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr13_+_18663600 | 0.36 |
ENSDART00000141647
|
selenou1a
|
selenoprotein U 1a |
| chr22_+_26443235 | 0.36 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr23_+_6232895 | 0.35 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr5_-_30487822 | 0.35 |
ENSDART00000189288
ENSDART00000183201 |
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of thraa+thrab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:1901827 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.4 | 1.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.4 | 1.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.4 | 1.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.4 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 1.4 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.3 | 45.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.3 | 1.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 1.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.3 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 1.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.7 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.2 | 0.8 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.7 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.2 | 1.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 1.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 1.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.1 | 0.4 | GO:1901296 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.4 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 2.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.5 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 0.2 | GO:0090190 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.5 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 1.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.3 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.3 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.3 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.6 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 1.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.1 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.8 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 1.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.6 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.4 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 2.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.4 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 1.0 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 1.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.8 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.1 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 2.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0061036 | positive regulation of chondrocyte differentiation(GO:0032332) positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 2.5 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.2 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.8 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.8 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.4 | GO:0046058 | cAMP biosynthetic process(GO:0006171) cAMP metabolic process(GO:0046058) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 2.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.5 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 45.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 1.9 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.2 | 1.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 1.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 2.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.2 | 1.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.6 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 2.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.2 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 3.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 8.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |