Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
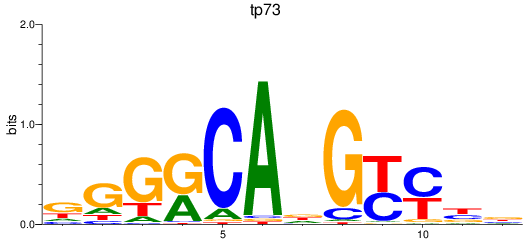
Results for tp73
Z-value: 0.89
Transcription factors associated with tp73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tp73
|
ENSDARG00000017953 | tumor protein p73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tp73 | dr11_v1_chr8_+_48848200_48848200 | 0.19 | 7.6e-01 | Click! |
Activity profile of tp73 motif
Sorted Z-values of tp73 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_12176327 | 0.72 |
ENSDART00000041914
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr3_+_31680592 | 0.69 |
ENSDART00000172456
|
mylk5
|
myosin, light chain kinase 5 |
| chr3_+_8825382 | 0.63 |
ENSDART00000158893
|
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr13_-_4707018 | 0.61 |
ENSDART00000128422
|
oit3
|
oncoprotein induced transcript 3 |
| chr21_+_26612777 | 0.61 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr19_+_4068134 | 0.59 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr2_+_36691771 | 0.58 |
ENSDART00000131978
|
ptx3b
|
pentraxin 3, long b |
| chr11_+_14287427 | 0.54 |
ENSDART00000180903
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr11_+_14333441 | 0.53 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr3_+_16229911 | 0.53 |
ENSDART00000121728
|
rpl19
|
ribosomal protein L19 |
| chr11_+_12175162 | 0.53 |
ENSDART00000125446
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr18_-_38270077 | 0.53 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr21_-_27338639 | 0.52 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr3_-_39488639 | 0.50 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr20_-_30376433 | 0.49 |
ENSDART00000190737
|
rps7
|
ribosomal protein S7 |
| chr5_+_40837539 | 0.48 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr3_-_39488482 | 0.48 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr8_+_22485956 | 0.47 |
ENSDART00000099960
|
si:ch211-261n11.7
|
si:ch211-261n11.7 |
| chr19_+_4061699 | 0.47 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr12_+_22576404 | 0.46 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr21_-_20832482 | 0.42 |
ENSDART00000191928
|
c6
|
complement component 6 |
| chr13_-_17464362 | 0.41 |
ENSDART00000145499
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr25_+_34641536 | 0.40 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr13_-_17464654 | 0.39 |
ENSDART00000140312
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr25_+_36292465 | 0.36 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr14_+_15484544 | 0.35 |
ENSDART00000188649
|
CR382326.1
|
|
| chr4_-_49607096 | 0.34 |
ENSDART00000154381
|
si:dkey-159n16.4
|
si:dkey-159n16.4 |
| chr11_+_30303785 | 0.34 |
ENSDART00000185610
ENSDART00000126815 |
ugt1b5
|
UDP glucuronosyltransferase 1 family, polypeptide B5 |
| chr5_-_26936861 | 0.34 |
ENSDART00000191032
|
htra4
|
HtrA serine peptidase 4 |
| chr7_-_16596938 | 0.31 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr1_-_53714885 | 0.29 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr18_+_20567542 | 0.29 |
ENSDART00000182585
|
bida
|
BH3 interacting domain death agonist |
| chr7_-_16597130 | 0.28 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr7_+_41314862 | 0.28 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr3_+_62219158 | 0.27 |
ENSDART00000162354
|
BX470259.5
|
|
| chr8_+_30747940 | 0.27 |
ENSDART00000098975
|
p2rx4b
|
purinergic receptor P2X, ligand-gated ion channel, 4b |
| chr8_+_25767610 | 0.25 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr7_-_73848458 | 0.25 |
ENSDART00000041385
|
zgc:163061
|
zgc:163061 |
| chr5_+_27898226 | 0.24 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
| chr6_+_13046720 | 0.24 |
ENSDART00000165896
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr11_+_24298432 | 0.23 |
ENSDART00000138487
|
si:dkey-76p14.2
|
si:dkey-76p14.2 |
| chr24_+_14859238 | 0.23 |
ENSDART00000113859
ENSDART00000138227 |
crispld1a
|
cysteine-rich secretory protein LCCL domain containing 1a |
| chr1_-_56363108 | 0.23 |
ENSDART00000021878
|
gcgrb
|
glucagon receptor b |
| chr6_-_2154137 | 0.22 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr20_-_1314355 | 0.21 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr8_-_34762163 | 0.21 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr14_-_16328331 | 0.21 |
ENSDART00000111262
|
pcdh12
|
protocadherin 12 |
| chr13_-_50546634 | 0.20 |
ENSDART00000192127
|
CU570781.2
|
|
| chr5_+_6854498 | 0.20 |
ENSDART00000148663
|
elac1
|
elaC ribonuclease Z 1 |
| chr10_+_2587234 | 0.20 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr11_+_16137302 | 0.19 |
ENSDART00000164892
|
pimr204
|
Pim proto-oncogene, serine/threonine kinase, related 204 |
| chr7_+_19495905 | 0.19 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr1_+_11711503 | 0.19 |
ENSDART00000138355
|
si:dkey-26i13.8
|
si:dkey-26i13.8 |
| chr2_+_44528255 | 0.19 |
ENSDART00000193834
|
pask
|
PAS domain containing serine/threonine kinase |
| chr20_-_13673566 | 0.18 |
ENSDART00000181641
|
TAGAP
|
si:ch211-122h15.4 |
| chr6_-_10902916 | 0.17 |
ENSDART00000122221
|
nfe2l2b
|
nuclear factor, erythroid 2-like 2b |
| chr11_-_8271374 | 0.17 |
ENSDART00000168253
|
pimr202
|
Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr7_+_19495379 | 0.17 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr15_-_35960250 | 0.16 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr12_-_44151296 | 0.16 |
ENSDART00000168734
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr1_+_55600504 | 0.15 |
ENSDART00000123946
|
umod
|
uromodulin |
| chr4_+_25680480 | 0.14 |
ENSDART00000100737
|
acot17
|
acyl-CoA thioesterase 17 |
| chr16_-_19568795 | 0.14 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr2_+_10771787 | 0.14 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr25_+_4581214 | 0.14 |
ENSDART00000185552
|
CABZ01068600.1
|
|
| chr15_-_4485828 | 0.14 |
ENSDART00000062868
|
tfdp2
|
transcription factor Dp-2 |
| chr22_+_12477996 | 0.13 |
ENSDART00000177704
|
CR847870.3
|
|
| chr23_-_20345473 | 0.13 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr18_+_44768829 | 0.13 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr12_+_26608883 | 0.11 |
ENSDART00000153094
|
si:dkey-148d16.5
|
si:dkey-148d16.5 |
| chr18_+_32916351 | 0.11 |
ENSDART00000170387
ENSDART00000161553 |
olfcg2
|
olfactory receptor C family, g2 |
| chr20_+_4157815 | 0.11 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr2_-_48539673 | 0.10 |
ENSDART00000168202
|
CR391991.4
|
|
| chr20_-_164300 | 0.08 |
ENSDART00000183354
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr1_+_54773877 | 0.07 |
ENSDART00000129831
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr4_+_71018579 | 0.06 |
ENSDART00000186727
|
si:dkeyp-80d11.10
|
si:dkeyp-80d11.10 |
| chr16_+_54829293 | 0.05 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr7_-_22941472 | 0.05 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr13_-_9300299 | 0.05 |
ENSDART00000144142
|
HTRA2 (1 of many)
|
si:dkey-33c12.12 |
| chr20_-_1314537 | 0.04 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr9_-_52847881 | 0.04 |
ENSDART00000166407
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr19_-_7525588 | 0.03 |
ENSDART00000092256
|
mrpl9
|
mitochondrial ribosomal protein L9 |
| chr4_+_25693463 | 0.03 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr6_-_43769332 | 0.03 |
ENSDART00000128174
|
foxp1b
|
forkhead box P1b |
| chr3_+_52545400 | 0.02 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr8_+_52515188 | 0.02 |
ENSDART00000163668
|
si:ch1073-392o20.2
|
si:ch1073-392o20.2 |
| chr22_+_30331414 | 0.01 |
ENSDART00000133482
|
BX649448.3
|
|
| chr22_+_24318131 | 0.00 |
ENSDART00000187360
ENSDART00000165618 |
ccdc50
|
coiled-coil domain containing 50 |
| chr13_-_36418921 | 0.00 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr2_-_37837472 | 0.00 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr3_+_24577021 | 0.00 |
ENSDART00000158449
|
sp100.3
|
SP110 nuclear body protein, tandem duplicate 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tp73
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.2 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.1 | 0.4 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.2 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 0.1 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.5 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.6 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.1 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.0 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |