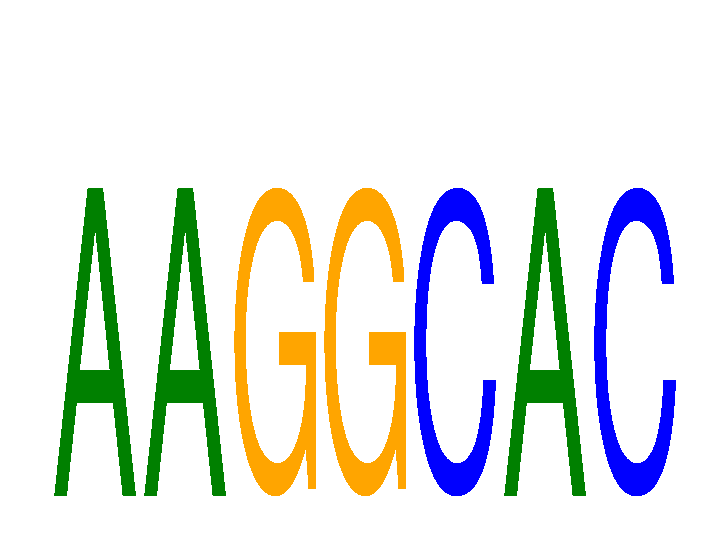Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AAGGCAC
Z-value: 1.82
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
mmu-miR-124-3p.1
|
MIMAT0000134 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_52181393 | 11.24 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr7_+_112278520 | 9.91 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr18_+_11052458 | 9.61 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr6_+_17307639 | 9.09 |
ENSMUST00000115453.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr2_+_35172392 | 8.89 |
ENSMUST00000028239.8
|
Gsn
|
gelsolin |
| chr16_+_24212284 | 8.86 |
ENSMUST00000038053.14
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_+_34818709 | 8.65 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr8_-_11362731 | 8.42 |
ENSMUST00000033898.10
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr16_+_78098344 | 8.23 |
ENSMUST00000232148.2
ENSMUST00000023572.15 |
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr7_+_89053562 | 8.18 |
ENSMUST00000058755.5
|
Fzd4
|
frizzled class receptor 4 |
| chr4_-_55532453 | 8.07 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr1_-_153208477 | 7.80 |
ENSMUST00000027752.15
|
Lamc1
|
laminin, gamma 1 |
| chr19_+_42135812 | 7.67 |
ENSMUST00000061111.10
|
Marveld1
|
MARVEL (membrane-associating) domain containing 1 |
| chr2_-_66615247 | 7.67 |
ENSMUST00000042792.7
|
Scn7a
|
sodium channel, voltage-gated, type VII, alpha |
| chr5_+_65127412 | 7.43 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr9_-_21223631 | 7.39 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr18_+_4921663 | 7.31 |
ENSMUST00000143254.8
|
Svil
|
supervillin |
| chrX_-_7537580 | 7.31 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr2_+_119156265 | 7.19 |
ENSMUST00000102517.4
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr13_-_60325170 | 7.09 |
ENSMUST00000065086.6
|
Gas1
|
growth arrest specific 1 |
| chr5_+_137348638 | 6.93 |
ENSMUST00000166239.8
ENSMUST00000111054.2 |
Ephb4
|
Eph receptor B4 |
| chr5_+_30745447 | 6.83 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr9_-_71499628 | 6.79 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr2_-_136958544 | 6.77 |
ENSMUST00000028735.8
|
Jag1
|
jagged 1 |
| chr14_-_22039543 | 6.77 |
ENSMUST00000043409.9
|
Zfp503
|
zinc finger protein 503 |
| chr5_+_32293145 | 6.51 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr2_+_27567213 | 6.46 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr10_-_62067026 | 6.32 |
ENSMUST00000047883.11
|
Tspan15
|
tetraspanin 15 |
| chr3_-_131065658 | 6.24 |
ENSMUST00000029610.9
|
Hadh
|
hydroxyacyl-Coenzyme A dehydrogenase |
| chr8_+_129412135 | 6.16 |
ENSMUST00000090006.12
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta) |
| chr1_+_74430575 | 5.98 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr15_-_102165884 | 5.98 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr18_+_58011691 | 5.97 |
ENSMUST00000115366.3
|
Slc12a2
|
solute carrier family 12, member 2 |
| chr15_-_89064936 | 5.90 |
ENSMUST00000109331.9
|
Plxnb2
|
plexin B2 |
| chr19_+_38384428 | 5.83 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr10_+_98943999 | 5.83 |
ENSMUST00000161240.4
|
Galnt4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr19_+_3373285 | 5.77 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr8_-_27664651 | 5.76 |
ENSMUST00000054212.7
ENSMUST00000033878.14 ENSMUST00000209377.2 |
Rab11fip1
|
RAB11 family interacting protein 1 (class I) |
| chr16_+_37597235 | 5.69 |
ENSMUST00000114763.3
|
Fstl1
|
follistatin-like 1 |
| chr1_-_134006847 | 5.61 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr5_-_98178811 | 5.55 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2 |
| chr6_-_120271520 | 5.48 |
ENSMUST00000057283.8
ENSMUST00000212457.2 |
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr19_-_31742427 | 5.47 |
ENSMUST00000065067.14
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr12_-_87435091 | 5.47 |
ENSMUST00000021424.5
|
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr7_-_98010534 | 5.45 |
ENSMUST00000165257.8
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr6_+_97783975 | 5.40 |
ENSMUST00000203884.3
ENSMUST00000043637.14 |
Mitf
|
melanogenesis associated transcription factor |
| chr3_-_85653573 | 5.38 |
ENSMUST00000118408.8
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chrX_-_141749704 | 5.38 |
ENSMUST00000041317.3
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr10_+_108167973 | 5.36 |
ENSMUST00000095313.5
|
Pawr
|
PRKC, apoptosis, WT1, regulator |
| chr7_+_88079465 | 5.28 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr13_+_5911481 | 5.27 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr10_-_7656635 | 5.27 |
ENSMUST00000124838.2
ENSMUST00000039763.14 |
Ginm1
|
glycoprotein integral membrane 1 |
| chr17_-_66258110 | 5.18 |
ENSMUST00000233580.2
ENSMUST00000024906.6 |
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr13_+_74269554 | 5.17 |
ENSMUST00000036208.7
ENSMUST00000225423.2 ENSMUST00000221703.2 |
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr17_+_64907697 | 5.16 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr1_-_64776890 | 5.15 |
ENSMUST00000116133.4
ENSMUST00000063982.7 |
Fzd5
|
frizzled class receptor 5 |
| chr11_+_75541324 | 5.11 |
ENSMUST00000102505.10
|
Myo1c
|
myosin IC |
| chr17_+_87270504 | 5.07 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr7_+_141047298 | 5.05 |
ENSMUST00000106000.10
ENSMUST00000209892.2 ENSMUST00000177840.9 |
Cd151
|
CD151 antigen |
| chr11_-_47270201 | 5.01 |
ENSMUST00000077221.6
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr2_+_32766126 | 4.99 |
ENSMUST00000028135.15
|
Niban2
|
niban apoptosis regulator 2 |
| chr7_+_44667377 | 4.98 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr11_+_53991750 | 4.98 |
ENSMUST00000093107.12
ENSMUST00000019050.12 ENSMUST00000174616.8 ENSMUST00000129499.8 ENSMUST00000126840.8 |
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr3_-_89294430 | 4.98 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr7_+_3341597 | 4.97 |
ENSMUST00000164553.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr17_+_5891582 | 4.97 |
ENSMUST00000002436.11
|
Snx9
|
sorting nexin 9 |
| chr19_+_27194757 | 4.96 |
ENSMUST00000047645.13
ENSMUST00000167487.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr13_-_113755082 | 4.94 |
ENSMUST00000109241.5
|
Snx18
|
sorting nexin 18 |
| chr16_+_14523696 | 4.92 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr2_+_31840151 | 4.85 |
ENSMUST00000001920.13
ENSMUST00000151276.3 |
Aif1l
|
allograft inflammatory factor 1-like |
| chr4_-_104967032 | 4.84 |
ENSMUST00000030243.8
|
Prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr2_+_165497157 | 4.83 |
ENSMUST00000063433.8
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr8_+_27575611 | 4.80 |
ENSMUST00000178514.8
ENSMUST00000033876.14 |
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr3_+_121220146 | 4.80 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr9_+_118755521 | 4.79 |
ENSMUST00000073109.12
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr7_-_132725075 | 4.76 |
ENSMUST00000163601.8
ENSMUST00000033269.15 ENSMUST00000124096.8 |
Ctbp2
Fgfr2
|
C-terminal binding protein 2 fibroblast growth factor receptor 2 |
| chr7_-_28661751 | 4.71 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr2_-_84481058 | 4.69 |
ENSMUST00000111670.9
ENSMUST00000111697.9 ENSMUST00000111696.8 ENSMUST00000111678.8 ENSMUST00000111690.8 ENSMUST00000111695.8 ENSMUST00000111677.8 ENSMUST00000111698.8 ENSMUST00000099941.9 ENSMUST00000111676.8 ENSMUST00000111694.8 ENSMUST00000111675.8 ENSMUST00000111689.8 ENSMUST00000111687.8 ENSMUST00000111692.8 ENSMUST00000111685.8 ENSMUST00000111686.8 ENSMUST00000111688.8 ENSMUST00000111693.8 ENSMUST00000111684.8 |
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr3_+_89325750 | 4.67 |
ENSMUST00000039110.12
ENSMUST00000125036.8 ENSMUST00000191485.7 ENSMUST00000154791.8 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr3_-_102111892 | 4.60 |
ENSMUST00000029453.13
|
Vangl1
|
VANGL planar cell polarity 1 |
| chr11_-_101357046 | 4.60 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chr2_+_13578738 | 4.59 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr5_+_145051025 | 4.54 |
ENSMUST00000085679.13
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr6_-_50359797 | 4.52 |
ENSMUST00000114468.9
|
Osbpl3
|
oxysterol binding protein-like 3 |
| chr14_-_47426863 | 4.51 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr18_-_63520195 | 4.51 |
ENSMUST00000047480.13
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr6_-_127128007 | 4.50 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr16_-_45664664 | 4.50 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr5_+_88868714 | 4.49 |
ENSMUST00000113229.8
ENSMUST00000006424.8 |
Mob1b
|
MOB kinase activator 1B |
| chr12_+_33003882 | 4.49 |
ENSMUST00000076698.13
|
Sypl
|
synaptophysin-like protein |
| chrX_+_98179725 | 4.49 |
ENSMUST00000052839.7
|
Efnb1
|
ephrin B1 |
| chr2_-_150510116 | 4.48 |
ENSMUST00000028944.4
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr13_-_111945499 | 4.47 |
ENSMUST00000109267.9
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr10_-_93146937 | 4.45 |
ENSMUST00000008542.12
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr17_-_84495364 | 4.45 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr15_+_88746380 | 4.44 |
ENSMUST00000042818.11
|
Pim3
|
proviral integration site 3 |
| chr6_-_92920466 | 4.44 |
ENSMUST00000113438.8
|
Adamts9
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 |
| chr4_-_11007635 | 4.44 |
ENSMUST00000054776.4
|
Plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr2_-_73360069 | 4.44 |
ENSMUST00000094681.11
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_-_84489783 | 4.44 |
ENSMUST00000107687.9
ENSMUST00000098990.10 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr14_-_118943591 | 4.42 |
ENSMUST00000036554.14
ENSMUST00000166646.2 |
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr19_-_47452840 | 4.40 |
ENSMUST00000081619.10
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr2_-_24825160 | 4.39 |
ENSMUST00000102935.10
ENSMUST00000133934.2 ENSMUST00000028349.14 |
Arrdc1
|
arrestin domain containing 1 |
| chr16_+_4412546 | 4.37 |
ENSMUST00000014447.13
|
Glis2
|
GLIS family zinc finger 2 |
| chr12_-_54842488 | 4.36 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chr15_-_77726333 | 4.36 |
ENSMUST00000016771.13
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr2_+_16361081 | 4.35 |
ENSMUST00000028081.13
|
Plxdc2
|
plexin domain containing 2 |
| chr19_+_32597379 | 4.33 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr9_+_72952115 | 4.32 |
ENSMUST00000184146.8
ENSMUST00000034722.5 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr7_-_80052491 | 4.28 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr6_-_38853097 | 4.26 |
ENSMUST00000161779.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr10_-_121312212 | 4.25 |
ENSMUST00000026902.9
|
Rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr19_+_16413126 | 4.25 |
ENSMUST00000025602.4
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chr8_+_93687561 | 4.24 |
ENSMUST00000072939.8
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr4_-_59549314 | 4.22 |
ENSMUST00000148331.9
ENSMUST00000030076.12 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr8_+_129085719 | 4.21 |
ENSMUST00000026917.10
|
Nrp1
|
neuropilin 1 |
| chr7_-_80453033 | 4.21 |
ENSMUST00000167377.3
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr5_-_114829032 | 4.20 |
ENSMUST00000012028.14
|
Gltp
|
glycolipid transfer protein |
| chrX_+_138511360 | 4.19 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr3_+_101993731 | 4.15 |
ENSMUST00000029454.12
|
Casq2
|
calsequestrin 2 |
| chr5_-_30360113 | 4.12 |
ENSMUST00000156859.3
|
Hadha
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| chrX_+_105070907 | 4.11 |
ENSMUST00000055941.7
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr3_-_19365431 | 4.10 |
ENSMUST00000099195.10
|
Pde7a
|
phosphodiesterase 7A |
| chr18_-_66135683 | 4.04 |
ENSMUST00000120461.9
ENSMUST00000048260.15 ENSMUST00000236866.2 |
Lman1
|
lectin, mannose-binding, 1 |
| chr3_+_37694094 | 4.04 |
ENSMUST00000108109.8
ENSMUST00000038569.8 ENSMUST00000108107.2 |
Spry1
|
sprouty RTK signaling antagonist 1 |
| chr6_+_42222841 | 4.03 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr11_-_76737794 | 4.01 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr2_+_162773440 | 4.00 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr3_+_89427458 | 3.99 |
ENSMUST00000000811.8
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr19_+_25214322 | 3.99 |
ENSMUST00000049400.15
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr18_+_75953244 | 3.98 |
ENSMUST00000058997.15
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr8_+_107662352 | 3.96 |
ENSMUST00000212524.2
ENSMUST00000047425.5 |
Sntb2
|
syntrophin, basic 2 |
| chr2_-_142743438 | 3.92 |
ENSMUST00000230763.2
ENSMUST00000043589.8 |
Kif16b
|
kinesin family member 16B |
| chr10_+_58230183 | 3.92 |
ENSMUST00000020077.11
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_-_34345014 | 3.91 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr5_+_137015873 | 3.87 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr6_-_119394634 | 3.87 |
ENSMUST00000032272.13
|
Adipor2
|
adiponectin receptor 2 |
| chr2_-_173118315 | 3.86 |
ENSMUST00000036248.13
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_-_21149392 | 3.84 |
ENSMUST00000037998.6
|
Tram2
|
translocating chain-associating membrane protein 2 |
| chr4_+_137589548 | 3.82 |
ENSMUST00000102518.10
|
Ece1
|
endothelin converting enzyme 1 |
| chr3_-_108053396 | 3.80 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr2_-_104647041 | 3.80 |
ENSMUST00000117237.2
ENSMUST00000231375.2 |
Qser1
|
glutamine and serine rich 1 |
| chr19_-_55087849 | 3.79 |
ENSMUST00000061856.6
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr8_-_62355690 | 3.79 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr10_+_59159118 | 3.79 |
ENSMUST00000009789.15
ENSMUST00000092512.11 ENSMUST00000105466.3 |
P4ha1
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
| chr11_+_49684984 | 3.78 |
ENSMUST00000020629.5
|
Gfpt2
|
glutamine fructose-6-phosphate transaminase 2 |
| chr5_-_147013384 | 3.76 |
ENSMUST00000016664.8
|
Lnx2
|
ligand of numb-protein X 2 |
| chr13_+_38529062 | 3.75 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr13_-_103911092 | 3.75 |
ENSMUST00000074616.7
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr6_+_38410848 | 3.75 |
ENSMUST00000160583.8
|
Ubn2
|
ubinuclein 2 |
| chr9_+_77824646 | 3.72 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr14_+_56091454 | 3.70 |
ENSMUST00000227465.2
ENSMUST00000168479.3 ENSMUST00000100529.10 |
Nynrin
|
NYN domain and retroviral integrase containing |
| chr5_+_64250268 | 3.68 |
ENSMUST00000087324.7
|
Pgm2
|
phosphoglucomutase 2 |
| chr18_+_84106188 | 3.68 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr8_-_127201801 | 3.67 |
ENSMUST00000170518.3
|
Tarbp1
|
TAR RNA binding protein 1 |
| chr2_-_152185901 | 3.66 |
ENSMUST00000040312.7
|
Trib3
|
tribbles pseudokinase 3 |
| chr3_-_87081939 | 3.66 |
ENSMUST00000159976.8
ENSMUST00000107618.9 |
Kirrel
|
kirre like nephrin family adhesion molecule 1 |
| chr4_-_40853950 | 3.65 |
ENSMUST00000030121.13
ENSMUST00000108096.3 |
B4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr2_-_118380149 | 3.61 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr9_-_31824758 | 3.60 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chrX_-_51254129 | 3.58 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr10_+_21870565 | 3.57 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_+_129181407 | 3.54 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr4_+_62204678 | 3.54 |
ENSMUST00000084530.9
|
Slc31a2
|
solute carrier family 31, member 2 |
| chr11_+_55360502 | 3.53 |
ENSMUST00000018727.4
|
G3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr7_-_65020955 | 3.52 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr12_+_111005768 | 3.48 |
ENSMUST00000084968.14
|
Rcor1
|
REST corepressor 1 |
| chr10_+_79690452 | 3.48 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr11_+_44508137 | 3.46 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr19_-_6065872 | 3.44 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr15_+_25622611 | 3.44 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr1_-_191129223 | 3.44 |
ENSMUST00000067976.9
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr6_-_39702381 | 3.43 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene |
| chr5_-_66775979 | 3.43 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr10_-_13350106 | 3.43 |
ENSMUST00000105545.12
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr5_+_73071607 | 3.42 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr2_-_60793536 | 3.42 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_-_155306992 | 3.41 |
ENSMUST00000084103.10
ENSMUST00000030917.6 |
Ski
|
ski sarcoma viral oncogene homolog (avian) |
| chr15_-_73056713 | 3.41 |
ENSMUST00000044113.12
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr11_+_118913788 | 3.38 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr12_+_116244792 | 3.37 |
ENSMUST00000100986.4
ENSMUST00000220816.2 |
Esyt2
|
extended synaptotagmin-like protein 2 |
| chr12_+_80837284 | 3.36 |
ENSMUST00000220238.2
ENSMUST00000068519.7 |
Susd6
|
sushi domain containing 6 |
| chr3_-_131138541 | 3.33 |
ENSMUST00000090246.5
ENSMUST00000126569.2 |
Sgms2
|
sphingomyelin synthase 2 |
| chr12_+_112112621 | 3.32 |
ENSMUST00000128402.3
|
Kif26a
|
kinesin family member 26A |
| chr3_+_122522592 | 3.32 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chrX_-_105055486 | 3.32 |
ENSMUST00000238718.2
ENSMUST00000033583.14 ENSMUST00000151689.9 |
Magt1
|
magnesium transporter 1 |
| chr1_-_120198804 | 3.30 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr10_-_60983438 | 3.28 |
ENSMUST00000092498.12
ENSMUST00000137833.2 ENSMUST00000155919.8 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr18_+_84869456 | 3.25 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr1_-_98023321 | 3.22 |
ENSMUST00000058762.15
ENSMUST00000097625.10 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr18_-_73836810 | 3.22 |
ENSMUST00000025393.14
|
Smad4
|
SMAD family member 4 |
| chr4_-_56947411 | 3.22 |
ENSMUST00000107609.4
ENSMUST00000068792.13 |
Tmem245
|
transmembrane protein 245 |
| chr11_+_87959067 | 3.22 |
ENSMUST00000018521.11
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr7_-_121620366 | 3.21 |
ENSMUST00000033160.15
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr15_+_102314809 | 3.21 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chr16_-_10810964 | 3.21 |
ENSMUST00000023143.14
|
Litaf
|
LPS-induced TN factor |
| chr8_+_35842872 | 3.18 |
ENSMUST00000210337.2
ENSMUST00000070481.8 ENSMUST00000211648.2 |
Ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr7_+_45084257 | 3.17 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr17_-_74601769 | 3.16 |
ENSMUST00000078459.8
ENSMUST00000232989.2 |
Memo1
|
mediator of cell motility 1 |
| chr6_-_137626207 | 3.16 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:0060574 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 3.3 | 13.3 | GO:1903921 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 2.9 | 8.6 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 2.7 | 8.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 2.4 | 7.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 2.3 | 7.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 2.3 | 13.9 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 2.3 | 6.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 2.1 | 6.2 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 2.0 | 12.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 2.0 | 8.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 1.9 | 7.6 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 1.8 | 5.4 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 1.7 | 5.2 | GO:0060715 | Spemann organizer formation(GO:0060061) syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 1.6 | 4.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 1.6 | 3.2 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 1.6 | 4.8 | GO:0060915 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 1.6 | 3.1 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 1.5 | 9.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.5 | 3.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 1.5 | 4.5 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 1.5 | 6.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.5 | 4.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 1.5 | 4.4 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 1.4 | 4.3 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 1.4 | 4.1 | GO:1904959 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.4 | 6.8 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.3 | 9.4 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.3 | 4.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.3 | 5.2 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 1.3 | 6.5 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 1.3 | 3.8 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.2 | 5.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.2 | 5.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 1.2 | 3.7 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 1.2 | 3.6 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 1.2 | 6.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) Harderian gland development(GO:0070384) |
| 1.2 | 11.9 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 1.2 | 1.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 1.2 | 4.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 1.1 | 4.5 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 1.1 | 2.2 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 1.1 | 5.5 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 1.1 | 4.3 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.1 | 3.2 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 1.1 | 8.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 1.1 | 3.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.0 | 3.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 1.0 | 3.0 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.0 | 8.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 1.0 | 7.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.0 | 2.9 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.9 | 0.9 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.9 | 1.8 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.9 | 0.9 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.9 | 3.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.8 | 5.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.8 | 2.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.8 | 2.5 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.8 | 3.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.8 | 5.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.8 | 2.4 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.8 | 12.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.8 | 2.4 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.8 | 4.7 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.8 | 4.7 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.8 | 3.9 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.8 | 4.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.8 | 1.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.8 | 5.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.8 | 2.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.8 | 3.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.8 | 5.3 | GO:0090383 | phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 0.7 | 5.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.7 | 2.9 | GO:0036233 | glycine import(GO:0036233) |
| 0.7 | 0.7 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.7 | 2.8 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.7 | 4.9 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.7 | 0.7 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.7 | 2.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.7 | 4.8 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.7 | 4.8 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.7 | 4.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.7 | 3.3 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.7 | 2.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.7 | 0.7 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.7 | 5.9 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.6 | 3.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.6 | 5.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.6 | 2.5 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.6 | 3.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.6 | 5.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.6 | 12.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.6 | 3.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.6 | 2.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.6 | 1.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.6 | 2.4 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.6 | 4.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.6 | 1.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.6 | 9.4 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.6 | 2.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.6 | 2.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.6 | 1.1 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.6 | 1.7 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.6 | 2.3 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.6 | 3.3 | GO:0060282 | positive regulation of chronic inflammatory response(GO:0002678) positive regulation of oocyte development(GO:0060282) |
| 0.5 | 1.6 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 1.6 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.5 | 3.8 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.5 | 9.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.5 | 3.8 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.5 | 2.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.5 | 4.8 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.5 | 1.6 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.5 | 2.7 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.5 | 11.6 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.5 | 4.2 | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.5 | 2.6 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.5 | 2.6 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.5 | 9.9 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.5 | 5.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.5 | 3.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.5 | 2.0 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.5 | 4.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.5 | 3.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.5 | 3.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.5 | 1.5 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.5 | 3.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.5 | 1.9 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.5 | 5.7 | GO:0050932 | positive regulation of developmental pigmentation(GO:0048087) regulation of pigment cell differentiation(GO:0050932) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.5 | 1.4 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.5 | 1.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.5 | 0.5 | GO:0010643 | atrial ventricular junction remodeling(GO:0003294) cell communication by chemical coupling(GO:0010643) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.5 | 2.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.5 | 1.8 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.4 | 3.6 | GO:0042637 | catagen(GO:0042637) |
| 0.4 | 2.7 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.4 | 4.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.4 | 1.3 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.4 | 5.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 2.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.4 | 2.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.4 | 2.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.4 | 1.7 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.4 | 2.5 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.4 | 3.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.4 | 2.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.4 | 5.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.4 | 2.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.4 | 3.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.4 | 1.2 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.4 | 1.2 | GO:2000847 | negative regulation of cell adhesion mediated by integrin(GO:0033629) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.4 | 5.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.4 | 3.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.4 | 2.0 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.4 | 1.9 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.4 | 5.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.4 | 3.8 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.4 | 4.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 2.2 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.4 | 2.6 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 1.8 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.4 | 1.8 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 | 3.9 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.4 | 3.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.4 | 2.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.3 | 3.1 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.3 | 5.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.3 | 1.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.3 | 3.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 4.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 1.3 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.3 | 2.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.3 | 1.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 2.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 2.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 7.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 2.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.3 | 1.9 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 2.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 0.6 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.3 | 0.9 | GO:0046021 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.3 | 1.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 2.5 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.3 | 3.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 3.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 0.9 | GO:0097156 | fasciculation of sensory neuron axon(GO:0097155) fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 0.9 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.3 | 4.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.3 | 2.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 2.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.3 | 1.5 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 2.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.3 | 3.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 1.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 1.5 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 0.9 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.3 | 5.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.3 | 1.1 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 3.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) negative regulation of fibroblast migration(GO:0010764) |
| 0.3 | 4.0 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 4.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.3 | 3.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 4.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 4.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.3 | 3.1 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 3.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.3 | 1.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 1.9 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 1.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.3 | 4.5 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.3 | 2.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.3 | 12.7 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.3 | 2.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.5 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.2 | 4.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 2.7 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.2 | 1.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 4.4 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.2 | 0.7 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) epicardium morphogenesis(GO:1905223) |
| 0.2 | 1.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.7 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 0.7 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) |
| 0.2 | 2.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 2.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 4.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 3.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 2.4 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.2 | 2.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 2.8 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.2 | 2.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.2 | 4.9 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 | 0.9 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 3.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.9 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.7 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.2 | 2.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 15.2 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 2.2 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 | 2.6 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 2.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 1.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 6.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.2 | 0.6 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.8 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.2 | 10.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 3.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 9.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 1.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 1.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 1.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 2.6 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.2 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.8 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 3.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 0.8 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 2.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 4.2 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.2 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 1.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 1.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 1.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.2 | 1.4 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 2.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 2.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.2 | 1.4 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 1.4 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 | 0.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 1.7 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.2 | 2.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.5 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.2 | 2.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 3.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 0.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 1.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 2.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 2.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 0.3 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.2 | 0.5 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.2 | 1.8 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.2 | 0.8 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 3.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.2 | 1.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.5 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.2 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 3.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 0.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 1.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 2.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 7.3 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 2.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.3 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 4.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 2.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 2.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 4.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 4.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.5 | GO:2001107 | negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 4.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 4.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 4.0 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 2.3 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.6 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.4 | GO:0022012 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) frontal suture morphogenesis(GO:0060364) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 2.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.2 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 2.0 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 1.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.4 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.6 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 1.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 2.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 2.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 5.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 9.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 3.7 | GO:0007588 | excretion(GO:0007588) |
| 0.1 | 0.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 1.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 2.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 1.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 3.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 3.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 2.5 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 3.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 6.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 5.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 0.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.7 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.1 | 1.8 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 2.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 1.9 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 1.8 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.2 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 1.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 3.2 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 2.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.9 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.2 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 0.5 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.6 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 1.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.7 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 1.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 1.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 2.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.0 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 3.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 1.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.8 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.1 | GO:0010730 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 2.9 | GO:0043297 | apical junction assembly(GO:0043297) |
| 0.1 | 4.5 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.1 | 1.3 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 1.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 2.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 2.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.8 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.5 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.0 | 1.0 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 2.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 11.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 2.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 1.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 1.0 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 1.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 2.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 1.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 1.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 1.2 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 2.6 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 1.3 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 2.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.7 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.0 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 1.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 2.7 | 8.2 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 2.5 | 9.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.9 | 7.8 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 1.8 | 8.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.6 | 4.7 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.4 | 4.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.3 | 5.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.2 | 8.4 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 1.1 | 4.5 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 1.1 | 4.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.1 | 1.1 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 1.0 | 5.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.0 | 7.7 | GO:0097422 | tubular endosome(GO:0097422) |
| 1.0 | 8.6 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.9 | 3.7 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.9 | 8.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.8 | 8.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.8 | 3.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.7 | 4.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.7 | 2.7 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.7 | 5.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.6 | 3.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.6 | 6.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.6 | 10.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.6 | 1.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.6 | 8.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.6 | 4.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 2.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.5 | 4.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 1.6 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.5 | 6.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 3.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.5 | 2.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.4 | 1.3 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.4 | 5.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 4.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 3.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.4 | 3.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 5.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 1.8 | GO:0035363 | histone locus body(GO:0035363) |
| 0.3 | 1.7 | GO:0044308 | axonal spine(GO:0044308) |
| 0.3 | 11.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 4.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.3 | 14.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.3 | 2.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 4.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 2.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 4.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 3.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 5.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 1.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) lumenal side of membrane(GO:0098576) |
| 0.3 | 5.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 9.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 1.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 2.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 7.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 13.1 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 7.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 4.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 9.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 2.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 3.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 2.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 5.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 1.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 1.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 2.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 4.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 3.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 5.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 6.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 2.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 8.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 3.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 5.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 3.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 10.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 4.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 8.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 4.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 4.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 9.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 3.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 7.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 6.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 14.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 24.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 5.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 3.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 6.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.9 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 14.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.8 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.1 | 2.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 2.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 4.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 3.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 6.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 6.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 8.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 6.9 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 1.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 36.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 8.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 30.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 0.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 3.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 2.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 3.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 24.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 9.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 5.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 3.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.8 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 10.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 5.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 4.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 5.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 4.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.6 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 92.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0001940 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 6.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 2.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 30.3 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0000502 | proteasome complex(GO:0000502) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.9 | 5.8 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 1.8 | 5.3 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 1.7 | 8.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.7 | 5.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 1.7 | 5.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 1.6 | 8.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 1.6 | 6.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.5 | 9.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.5 | 8.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 1.4 | 4.2 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 1.4 | 4.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.2 | 6.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 1.2 | 9.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.2 | 3.7 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 1.2 | 3.7 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 1.1 | 3.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 1.1 | 4.3 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.1 | 3.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 1.0 | 13.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.0 | 6.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.0 | 4.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.0 | 3.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.9 | 12.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.9 | 3.7 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.9 | 5.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.9 | 5.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.9 | 4.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.8 | 8.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.8 | 5.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.8 | 7.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.8 | 2.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.8 | 2.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.8 | 3.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 4.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.8 | 4.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 3.9 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.8 | 3.9 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.8 | 16.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.8 | 3.8 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.8 | 6.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.7 | 4.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.7 | 2.9 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.7 | 2.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.7 | 2.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.7 | 4.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.7 | 2.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.6 | 2.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.6 | 1.8 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.6 | 13.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 1.8 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.6 | 2.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.6 | 5.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.5 | 1.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 1.6 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.5 | 9.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.5 | 2.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.5 | 7.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 8.7 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.5 | 5.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.5 | 4.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 3.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.5 | 8.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 1.5 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.5 | 2.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 1.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.5 | 2.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.5 | 4.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.5 | 6.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 3.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 1.8 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.4 | 3.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 1.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.4 | 4.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.4 | 6.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 3.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.4 | 2.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 4.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 3.3 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 7.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.4 | 13.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 14.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.4 | 3.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.4 | 2.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.4 | 1.6 | GO:0009041 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.4 | 1.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.4 | 2.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 3.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.4 | 4.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 3.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 4.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 4.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.4 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 1.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.4 | 6.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 1.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 4.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 2.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 1.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 5.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 4.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 2.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.3 | 7.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 4.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 1.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.3 | 2.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 3.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 1.9 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 3.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 1.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 16.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 3.7 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 3.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 2.7 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.3 | 6.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 1.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 0.9 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.3 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 1.5 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 4.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 0.9 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.3 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 9.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 5.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.3 | 7.3 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.3 | 3.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.3 | 4.2 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.3 | 1.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 2.6 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 2.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 7.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 6.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 7.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 3.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 2.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 2.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 4.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 1.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 2.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 1.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 2.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 0.6 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 2.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 1.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 3.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 0.7 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 5.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 4.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 2.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 3.7 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 3.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 1.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 2.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 2.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 3.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 2.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 2.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 2.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 1.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 3.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 4.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 3.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 3.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 4.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 3.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 3.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.7 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 3.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 3.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 4.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.6 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 1.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 3.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 2.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 3.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 31.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.7 | GO:0001093 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 2.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 4.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 5.1 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.1 | 3.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 4.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 4.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 3.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 15.8 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 0.4 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 3.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.1 | 1.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 3.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 16.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.5 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 2.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 7.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.1 | 2.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 3.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 2.8 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 0.9 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 1.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 34.8 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.1 | 6.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.9 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 5.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 1.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 6.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.7 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 1.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 3.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 5.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 10.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 3.1 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 2.4 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.6 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.2 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.9 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 3.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.7 | 21.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 10.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 20.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.5 | 18.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.5 | 3.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.3 | 10.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 27.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 12.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.3 | 13.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.3 | 11.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 18.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 5.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 12.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 8.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 9.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 13.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.3 | 12.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.3 | 10.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 2.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 7.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 3.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 8.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 18.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 11.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 25.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 6.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 12.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 10.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 5.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 3.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 14.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 7.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 8.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 2.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 9.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 6.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 7.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 4.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 23.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 14.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 5.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 4.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 4.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 5.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 5.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 23.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 16.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.7 | 12.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.7 | 7.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 17.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 22.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 6.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 6.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 8.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 17.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 4.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 11.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 8.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 4.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.4 | 10.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 5.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 12.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 6.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 4.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 7.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 4.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 7.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 4.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 9.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 6.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 7.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 2.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 5.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 16.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 6.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 12.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 4.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 15.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 4.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 3.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 11.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 5.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 4.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 4.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 4.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 5.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.2 | 4.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 3.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 4.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 2.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 2.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 5.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 3.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 2.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 3.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 8.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 10.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 4.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 3.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 4.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 4.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 5.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.8 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 4.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.8 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 0.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 3.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 14.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 12.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.9 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 8.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 5.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.3 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 7.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 3.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |