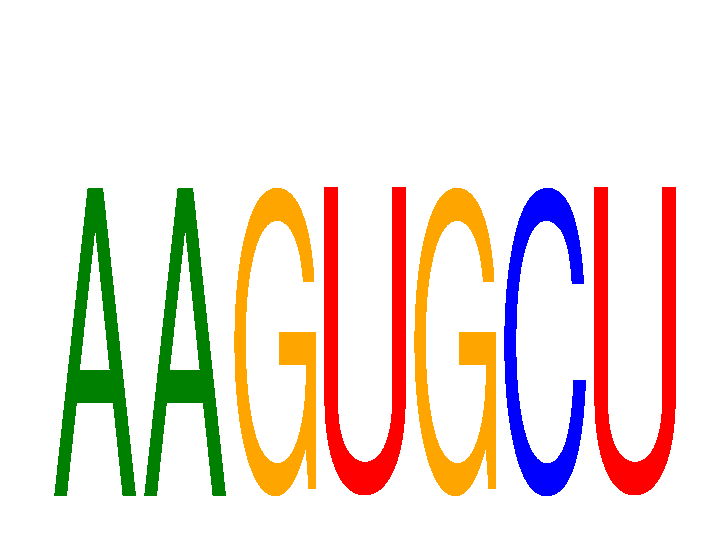Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AAGUGCU
Z-value: 0.65
miRNA associated with seed AAGUGCU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-291a-3p
|
MIMAT0000368 |
|
mmu-miR-294-3p
|
MIMAT0000372 |
|
mmu-miR-295-3p
|
MIMAT0000373 |
|
mmu-miR-302a-3p
|
MIMAT0000380 |
|
mmu-miR-302b-3p
|
MIMAT0003374 |
|
mmu-miR-302d-3p
|
MIMAT0003377 |
Activity profile of AAGUGCU motif
Sorted Z-values of AAGUGCU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGUGCU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_135899678 | 4.65 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr2_+_127178072 | 4.53 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr1_-_69724939 | 3.61 |
ENSMUST00000027146.9
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr16_+_57173456 | 3.51 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr4_+_11156411 | 3.33 |
ENSMUST00000029865.4
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1 |
| chr5_+_3393893 | 3.09 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr5_-_136596299 | 2.98 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chr13_-_111945499 | 2.97 |
ENSMUST00000109267.9
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr17_-_32074754 | 2.88 |
ENSMUST00000024839.6
|
Sik1
|
salt inducible kinase 1 |
| chr5_-_21156766 | 2.80 |
ENSMUST00000036489.10
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr2_-_131871725 | 2.74 |
ENSMUST00000028814.15
|
Rassf2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr16_-_23807602 | 2.68 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr5_+_77413282 | 2.67 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr5_+_64960705 | 2.58 |
ENSMUST00000165536.8
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr1_-_45964730 | 2.58 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr3_+_96465265 | 2.57 |
ENSMUST00000074519.13
ENSMUST00000049093.8 |
Txnip
|
thioredoxin interacting protein |
| chr11_-_103158190 | 2.56 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr10_+_96452860 | 2.37 |
ENSMUST00000038377.9
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr11_-_5211558 | 2.34 |
ENSMUST00000020662.15
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr12_-_118265103 | 2.22 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr9_-_44792575 | 2.22 |
ENSMUST00000114689.8
ENSMUST00000002095.11 ENSMUST00000128768.3 |
Kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr2_+_4405769 | 2.10 |
ENSMUST00000075767.14
|
Frmd4a
|
FERM domain containing 4A |
| chr3_-_27208010 | 2.09 |
ENSMUST00000108300.8
ENSMUST00000108298.9 |
Ect2
|
ect2 oncogene |
| chr13_+_111822712 | 2.08 |
ENSMUST00000109272.9
|
Mier3
|
MIER family member 3 |
| chr10_-_128540847 | 1.98 |
ENSMUST00000026415.9
ENSMUST00000026416.15 |
Cdk2
|
cyclin-dependent kinase 2 |
| chr12_+_71062733 | 1.97 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr5_-_44383943 | 1.96 |
ENSMUST00000055128.12
|
Tapt1
|
transmembrane anterior posterior transformation 1 |
| chr5_+_73071607 | 1.95 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr3_+_95566082 | 1.95 |
ENSMUST00000037947.15
ENSMUST00000178686.2 |
Mcl1
|
myeloid cell leukemia sequence 1 |
| chr12_-_107969853 | 1.91 |
ENSMUST00000066060.11
|
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr6_-_99243455 | 1.90 |
ENSMUST00000113326.9
|
Foxp1
|
forkhead box P1 |
| chr5_+_137517140 | 1.89 |
ENSMUST00000031727.10
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr3_+_60408600 | 1.86 |
ENSMUST00000099087.8
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr9_+_7764042 | 1.84 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr2_-_104647041 | 1.83 |
ENSMUST00000117237.2
ENSMUST00000231375.2 |
Qser1
|
glutamine and serine rich 1 |
| chrX_-_50294652 | 1.78 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr19_+_40819682 | 1.75 |
ENSMUST00000025983.13
ENSMUST00000119316.2 |
Ccnj
|
cyclin J |
| chr15_-_57998443 | 1.74 |
ENSMUST00000038194.5
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr4_-_126362372 | 1.73 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr2_-_153083322 | 1.72 |
ENSMUST00000056924.14
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr2_-_19002932 | 1.69 |
ENSMUST00000006912.12
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr8_-_127320685 | 1.68 |
ENSMUST00000054960.9
|
Irf2bp2
|
interferon regulatory factor 2 binding protein 2 |
| chr12_-_79343040 | 1.67 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr13_-_117162041 | 1.63 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_+_72306503 | 1.62 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr11_+_97554192 | 1.61 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr3_+_130904000 | 1.55 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr19_-_14575395 | 1.49 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr11_-_49603501 | 1.48 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr10_+_53473032 | 1.47 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr6_-_127128007 | 1.46 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr10_+_70080913 | 1.44 |
ENSMUST00000046807.7
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chr1_+_131935342 | 1.41 |
ENSMUST00000086556.12
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr2_-_156848923 | 1.41 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr9_-_116004265 | 1.38 |
ENSMUST00000061101.12
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr5_-_96309849 | 1.35 |
ENSMUST00000155901.8
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr8_-_35962641 | 1.34 |
ENSMUST00000033927.8
|
Eri1
|
exoribonuclease 1 |
| chr4_-_3938352 | 1.34 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr11_-_103303480 | 1.34 |
ENSMUST00000041272.10
|
Plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr4_+_97665843 | 1.33 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr16_+_36832119 | 1.32 |
ENSMUST00000071452.12
ENSMUST00000054034.7 |
Polq
|
polymerase (DNA directed), theta |
| chr10_-_128727542 | 1.32 |
ENSMUST00000026408.7
|
Gdf11
|
growth differentiation factor 11 |
| chr9_+_30941924 | 1.31 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr3_+_28317354 | 1.27 |
ENSMUST00000159236.9
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr5_+_147367237 | 1.27 |
ENSMUST00000176600.8
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr8_+_23349543 | 1.26 |
ENSMUST00000238975.2
ENSMUST00000110696.8 ENSMUST00000044331.7 |
Kat6a
|
K(lysine) acetyltransferase 6A |
| chr1_-_9770434 | 1.25 |
ENSMUST00000088658.11
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr19_+_41471067 | 1.24 |
ENSMUST00000067795.13
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr2_-_157046386 | 1.24 |
ENSMUST00000029170.8
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr6_+_91955820 | 1.22 |
ENSMUST00000089334.9
|
Fgd5
|
FYVE, RhoGEF and PH domain containing 5 |
| chr13_+_47196975 | 1.21 |
ENSMUST00000037025.16
ENSMUST00000143868.2 |
Kdm1b
|
lysine (K)-specific demethylase 1B |
| chr4_-_135749032 | 1.21 |
ENSMUST00000030427.6
|
Eloa
|
elongin A |
| chrX_+_158038778 | 1.20 |
ENSMUST00000126686.8
ENSMUST00000033671.13 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr7_-_63588610 | 1.20 |
ENSMUST00000063694.10
|
Klf13
|
Kruppel-like factor 13 |
| chr11_-_51891259 | 1.19 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr7_-_67022520 | 1.19 |
ENSMUST00000156690.8
ENSMUST00000107476.8 ENSMUST00000076325.12 ENSMUST00000032776.15 ENSMUST00000133074.2 |
Mef2a
|
myocyte enhancer factor 2A |
| chr15_-_66985760 | 1.18 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr8_+_85753452 | 1.18 |
ENSMUST00000047281.10
|
Trir
|
telomerase RNA component interacting RNase |
| chr9_+_121946321 | 1.16 |
ENSMUST00000119215.9
ENSMUST00000146832.8 ENSMUST00000118886.9 ENSMUST00000120173.9 ENSMUST00000139181.2 |
Snrk
|
SNF related kinase |
| chr11_-_46203047 | 1.15 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_+_65435825 | 1.14 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr6_-_83418656 | 1.13 |
ENSMUST00000089622.11
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr17_-_88372671 | 1.11 |
ENSMUST00000235112.2
ENSMUST00000005504.15 |
Fbxo11
|
F-box protein 11 |
| chr17_-_52139693 | 1.10 |
ENSMUST00000144331.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr14_+_103887644 | 1.09 |
ENSMUST00000069443.14
|
Slain1
|
SLAIN motif family, member 1 |
| chr11_-_115405200 | 1.09 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1 |
| chr18_+_3507917 | 1.05 |
ENSMUST00000025075.3
|
Bambi
|
BMP and activin membrane-bound inhibitor |
| chr18_+_89215543 | 1.05 |
ENSMUST00000037142.13
|
Cd226
|
CD226 antigen |
| chr15_+_95688712 | 1.05 |
ENSMUST00000071874.8
|
Ano6
|
anoctamin 6 |
| chr11_-_72302520 | 1.04 |
ENSMUST00000108500.8
ENSMUST00000050226.7 |
Smtnl2
|
smoothelin-like 2 |
| chr19_-_30526916 | 1.03 |
ENSMUST00000025803.9
|
Dkk1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr15_+_6737853 | 1.03 |
ENSMUST00000061656.8
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr17_-_90217868 | 1.01 |
ENSMUST00000086423.6
|
Gm10184
|
predicted pseudogene 10184 |
| chr1_+_6284823 | 1.00 |
ENSMUST00000027040.13
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr2_-_6889783 | 1.00 |
ENSMUST00000170438.8
ENSMUST00000114924.10 ENSMUST00000114934.11 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chrX_-_77627486 | 1.00 |
ENSMUST00000114025.8
ENSMUST00000134602.8 ENSMUST00000114024.9 |
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr15_+_100513230 | 0.99 |
ENSMUST00000000356.10
|
Dazap2
|
DAZ associated protein 2 |
| chr11_-_4544751 | 0.97 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr16_-_31898088 | 0.97 |
ENSMUST00000023467.9
|
Pak2
|
p21 (RAC1) activated kinase 2 |
| chr4_+_41135730 | 0.97 |
ENSMUST00000040008.4
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr6_+_120643323 | 0.94 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr1_-_74163575 | 0.94 |
ENSMUST00000169786.8
ENSMUST00000212888.2 ENSMUST00000191104.7 |
Tns1
|
tensin 1 |
| chr5_-_143717970 | 0.94 |
ENSMUST00000053287.6
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr10_+_42554888 | 0.94 |
ENSMUST00000040718.6
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr12_-_65010124 | 0.94 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr13_+_83652352 | 0.91 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_110581293 | 0.89 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr4_+_59805829 | 0.89 |
ENSMUST00000030080.7
|
Snx30
|
sorting nexin family member 30 |
| chr1_-_165287999 | 0.88 |
ENSMUST00000027856.13
|
Dcaf6
|
DDB1 and CUL4 associated factor 6 |
| chr16_-_92622972 | 0.88 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr13_+_45660905 | 0.88 |
ENSMUST00000000260.13
|
Gmpr
|
guanosine monophosphate reductase |
| chrX_-_47543029 | 0.88 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr4_-_133615075 | 0.87 |
ENSMUST00000003741.16
ENSMUST00000105894.11 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr14_-_103336990 | 0.86 |
ENSMUST00000022720.15
ENSMUST00000144141.8 |
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr6_+_140568366 | 0.85 |
ENSMUST00000032359.15
|
Aebp2
|
AE binding protein 2 |
| chr2_-_60383647 | 0.85 |
ENSMUST00000112525.5
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr1_-_180524587 | 0.83 |
ENSMUST00000027778.8
|
Mixl1
|
Mix1 homeobox-like 1 (Xenopus laevis) |
| chr4_-_86530245 | 0.83 |
ENSMUST00000125481.3
ENSMUST00000070607.9 |
Haus6
|
HAUS augmin-like complex, subunit 6 |
| chr4_+_32982981 | 0.82 |
ENSMUST00000098190.10
ENSMUST00000029946.14 |
Rragd
|
Ras-related GTP binding D |
| chr13_-_55477535 | 0.82 |
ENSMUST00000021941.8
|
Mxd3
|
Max dimerization protein 3 |
| chr9_-_59393893 | 0.82 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr11_-_104333059 | 0.80 |
ENSMUST00000106977.8
ENSMUST00000106972.8 |
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr11_-_34674677 | 0.80 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr5_-_25703700 | 0.80 |
ENSMUST00000173073.8
ENSMUST00000045291.14 ENSMUST00000173174.2 |
Kmt2c
|
lysine (K)-specific methyltransferase 2C |
| chr1_-_181039509 | 0.78 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr17_+_53873964 | 0.78 |
ENSMUST00000000724.15
|
Kat2b
|
K(lysine) acetyltransferase 2B |
| chr2_+_127967951 | 0.77 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr17_+_27192135 | 0.77 |
ENSMUST00000120016.3
|
Zbtb9
|
zinc finger and BTB domain containing 9 |
| chr8_-_86281946 | 0.76 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr18_+_32948436 | 0.76 |
ENSMUST00000025237.5
|
Tslp
|
thymic stromal lymphopoietin |
| chr19_+_53298906 | 0.75 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr18_-_46658957 | 0.75 |
ENSMUST00000036226.6
|
Fem1c
|
fem 1 homolog c |
| chr16_+_42727926 | 0.75 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_119765343 | 0.75 |
ENSMUST00000064091.12
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr6_-_90787100 | 0.74 |
ENSMUST00000101151.10
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr1_-_80318197 | 0.73 |
ENSMUST00000163119.8
|
Cul3
|
cullin 3 |
| chr17_-_48716756 | 0.71 |
ENSMUST00000160319.8
ENSMUST00000159535.2 ENSMUST00000078800.13 ENSMUST00000046719.14 ENSMUST00000162460.8 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr10_-_85752765 | 0.71 |
ENSMUST00000037646.9
ENSMUST00000220032.2 |
Prdm4
|
PR domain containing 4 |
| chr2_+_16361081 | 0.71 |
ENSMUST00000028081.13
|
Plxdc2
|
plexin domain containing 2 |
| chr4_+_106173384 | 0.70 |
ENSMUST00000165709.8
ENSMUST00000094933.5 |
Usp24
|
ubiquitin specific peptidase 24 |
| chr17_+_26934617 | 0.69 |
ENSMUST00000062519.14
ENSMUST00000144221.2 ENSMUST00000142539.8 ENSMUST00000151681.2 |
Crebrf
|
CREB3 regulatory factor |
| chr2_-_144112700 | 0.69 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5 |
| chr11_+_60428788 | 0.68 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr4_-_128699838 | 0.68 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr3_-_100396635 | 0.67 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr5_-_38719025 | 0.67 |
ENSMUST00000005234.13
|
Wdr1
|
WD repeat domain 1 |
| chr8_-_27664651 | 0.67 |
ENSMUST00000054212.7
ENSMUST00000033878.14 ENSMUST00000209377.2 |
Rab11fip1
|
RAB11 family interacting protein 1 (class I) |
| chr17_+_25407352 | 0.66 |
ENSMUST00000039734.12
|
Unkl
|
unkempt family like zinc finger |
| chr5_+_139366336 | 0.65 |
ENSMUST00000051293.8
|
Gpr146
|
G protein-coupled receptor 146 |
| chr13_+_43551851 | 0.64 |
ENSMUST00000071926.5
ENSMUST00000222499.2 |
Nol7
|
nucleolar protein 7 |
| chr3_-_144275897 | 0.63 |
ENSMUST00000043325.9
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr19_+_18648088 | 0.62 |
ENSMUST00000025632.11
|
Carnmt1
|
carnosine N-methyltransferase 1 |
| chr13_-_47083194 | 0.62 |
ENSMUST00000056978.8
|
Kif13a
|
kinesin family member 13A |
| chr4_-_44167509 | 0.62 |
ENSMUST00000098098.9
|
Rnf38
|
ring finger protein 38 |
| chr19_-_53378990 | 0.61 |
ENSMUST00000025997.7
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr13_-_9814407 | 0.61 |
ENSMUST00000146059.8
ENSMUST00000110637.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr1_-_119349969 | 0.61 |
ENSMUST00000038765.6
|
Inhbb
|
inhibin beta-B |
| chr12_-_100691251 | 0.60 |
ENSMUST00000043599.7
|
Rps6ka5
|
ribosomal protein S6 kinase, polypeptide 5 |
| chr18_-_46331877 | 0.59 |
ENSMUST00000167364.9
|
Trim36
|
tripartite motif-containing 36 |
| chr7_+_58878490 | 0.59 |
ENSMUST00000202945.4
ENSMUST00000107537.5 |
Ube3a
|
ubiquitin protein ligase E3A |
| chr19_+_3817396 | 0.59 |
ENSMUST00000052699.13
ENSMUST00000113974.11 ENSMUST00000113972.9 ENSMUST00000113973.8 ENSMUST00000113977.9 ENSMUST00000113968.9 |
Kmt5b
|
lysine methyltransferase 5B |
| chr19_+_46490123 | 0.59 |
ENSMUST00000026008.9
|
Trim8
|
tripartite motif-containing 8 |
| chr11_+_95304903 | 0.59 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr16_-_4377640 | 0.57 |
ENSMUST00000005862.9
|
Tfap4
|
transcription factor AP4 |
| chr11_+_77576981 | 0.57 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr19_+_44551280 | 0.56 |
ENSMUST00000040455.5
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr2_-_104324745 | 0.56 |
ENSMUST00000028600.14
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr6_-_84570890 | 0.55 |
ENSMUST00000168003.9
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr8_+_11808349 | 0.55 |
ENSMUST00000074856.13
ENSMUST00000098938.9 |
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr11_-_106890307 | 0.54 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr6_+_92068361 | 0.54 |
ENSMUST00000113460.8
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr11_-_65160767 | 0.54 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr18_+_70701260 | 0.53 |
ENSMUST00000074058.11
ENSMUST00000114946.4 |
Mbd2
|
methyl-CpG binding domain protein 2 |
| chr5_-_67585137 | 0.52 |
ENSMUST00000169190.5
|
Bend4
|
BEN domain containing 4 |
| chr6_+_141194886 | 0.52 |
ENSMUST00000043259.10
|
Pde3a
|
phosphodiesterase 3A, cGMP inhibited |
| chr7_+_144450260 | 0.51 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr2_-_33358513 | 0.51 |
ENSMUST00000113156.2
ENSMUST00000028125.12 ENSMUST00000126442.2 |
Zbtb43
|
zinc finger and BTB domain containing 43 |
| chr6_-_39396691 | 0.50 |
ENSMUST00000146785.8
ENSMUST00000114823.8 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr8_+_106786190 | 0.49 |
ENSMUST00000109308.3
|
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr16_+_33201164 | 0.49 |
ENSMUST00000165418.9
|
Zfp148
|
zinc finger protein 148 |
| chr14_-_57983511 | 0.48 |
ENSMUST00000173990.8
ENSMUST00000022531.14 |
Lats2
|
large tumor suppressor 2 |
| chr4_+_15265798 | 0.48 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr9_+_15217548 | 0.47 |
ENSMUST00000215124.2
ENSMUST00000164079.9 ENSMUST00000216109.2 |
Taf1d
|
TATA-box binding protein associated factor, RNA polymerase I, D |
| chr2_+_118731860 | 0.47 |
ENSMUST00000036578.7
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr3_+_121746862 | 0.45 |
ENSMUST00000037958.14
ENSMUST00000196904.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr15_-_64184485 | 0.45 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr3_+_88439616 | 0.45 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr9_-_114393406 | 0.45 |
ENSMUST00000111816.3
|
Trim71
|
tripartite motif-containing 71 |
| chr3_+_16237371 | 0.45 |
ENSMUST00000108345.9
ENSMUST00000191774.6 ENSMUST00000108346.5 |
Ythdf3
|
YTH N6-methyladenosine RNA binding protein 3 |
| chr19_+_25483070 | 0.45 |
ENSMUST00000087525.5
|
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr18_+_32296126 | 0.44 |
ENSMUST00000096575.5
|
Map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr16_-_97723753 | 0.44 |
ENSMUST00000170757.3
|
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr18_-_61533434 | 0.44 |
ENSMUST00000063307.6
ENSMUST00000075299.13 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr6_-_116605914 | 0.44 |
ENSMUST00000079749.6
|
Zfp422
|
zinc finger protein 422 |
| chr10_-_118705029 | 0.43 |
ENSMUST00000004281.10
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr11_+_4207557 | 0.42 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr13_+_14238361 | 0.42 |
ENSMUST00000129488.8
ENSMUST00000110536.8 ENSMUST00000110534.8 ENSMUST00000039538.15 ENSMUST00000110533.2 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr11_-_86884507 | 0.42 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr13_-_46118433 | 0.42 |
ENSMUST00000167708.4
ENSMUST00000091628.11 ENSMUST00000180110.9 |
Atxn1
|
ataxin 1 |
| chr11_+_104498824 | 0.42 |
ENSMUST00000021028.5
|
Itgb3
|
integrin beta 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 1.0 | 3.1 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.9 | 2.6 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.8 | 6.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.7 | 2.7 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.7 | 2.7 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.6 | 1.9 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.6 | 1.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.6 | 2.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.5 | 1.6 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.5 | 1.0 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.5 | 2.0 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.4 | 0.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.4 | 1.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.4 | 1.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.4 | 1.6 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.4 | 2.9 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.4 | 1.4 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.3 | 3.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 1.0 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.3 | 4.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 1.0 | GO:0061723 | glycophagy(GO:0061723) |
| 0.3 | 0.8 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.2 | 3.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 1.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.2 | 0.9 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 1.9 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 0.6 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.2 | 0.6 | GO:0035037 | sperm entry(GO:0035037) |
| 0.2 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 1.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.7 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 0.5 | GO:1904753 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.2 | 1.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 | 1.7 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 | 2.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.9 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 0.6 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.2 | 1.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.4 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 | 0.4 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 1.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.5 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.6 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.1 | 3.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 1.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.1 | 0.5 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 1.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.7 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.3 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 2.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.4 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 1.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 5.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 1.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.6 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.6 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.6 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.8 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 0.9 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.4 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 2.6 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.9 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 1.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.9 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 1.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 2.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.8 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 3.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 2.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 2.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 2.0 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 1.9 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 1.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.4 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.8 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 1.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 1.8 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 1.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.7 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 1.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 1.4 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.4 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.8 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.4 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.8 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.4 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 1.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 2.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0035561 | regulation of chromatin binding(GO:0035561) |
| 0.0 | 0.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.7 | 2.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.5 | 1.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 2.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.3 | 0.9 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 0.9 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 1.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.7 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 2.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 1.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 0.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.5 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.2 | 1.7 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 1.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.4 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) integrin alphav-beta3 complex(GO:0034683) |
| 0.1 | 0.8 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.1 | 0.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 3.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.6 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.3 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 3.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 3.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 6.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 2.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.6 | 1.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.5 | 3.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.5 | 1.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.4 | 2.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.4 | 2.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 1.3 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.3 | 3.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 1.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 0.9 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 4.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 1.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.7 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 4.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 2.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 2.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 2.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.7 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.0 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 3.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.8 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 3.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 2.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 2.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 2.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.6 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 1.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 3.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 2.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 5.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 9.8 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 4.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 4.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |