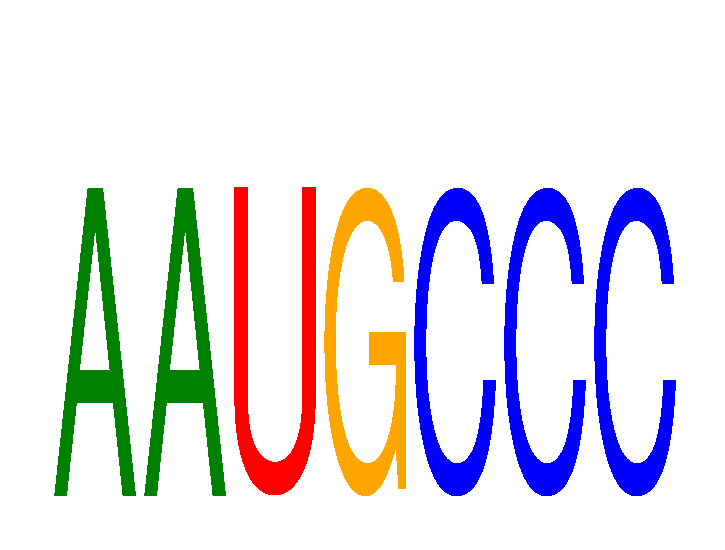Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AAUGCCC
Z-value: 0.57
miRNA associated with seed AAUGCCC
| Name | miRBASE accession |
|---|---|
|
mmu-miR-365-3p
|
MIMAT0000711 |
Activity profile of AAUGCCC motif
Sorted Z-values of AAUGCCC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAUGCCC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_103133524 | 4.63 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr16_+_34605282 | 3.94 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr5_+_64960705 | 3.51 |
ENSMUST00000165536.8
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr1_-_64776890 | 3.03 |
ENSMUST00000116133.4
ENSMUST00000063982.7 |
Fzd5
|
frizzled class receptor 5 |
| chr13_+_81031512 | 2.78 |
ENSMUST00000099356.10
|
Arrdc3
|
arrestin domain containing 3 |
| chr1_-_13660027 | 2.74 |
ENSMUST00000027068.11
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr6_-_52203146 | 2.64 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chrX_-_141749704 | 2.62 |
ENSMUST00000041317.3
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr18_-_61533434 | 2.60 |
ENSMUST00000063307.6
ENSMUST00000075299.13 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr11_-_53313950 | 2.50 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr10_+_98943999 | 2.28 |
ENSMUST00000161240.4
|
Galnt4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr7_-_118091135 | 2.26 |
ENSMUST00000178344.3
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
| chr8_-_80784619 | 2.12 |
ENSMUST00000079038.4
|
Hhip
|
Hedgehog-interacting protein |
| chr3_+_90434160 | 2.07 |
ENSMUST00000199538.5
ENSMUST00000164481.7 ENSMUST00000167598.6 |
S100a14
|
S100 calcium binding protein A14 |
| chr10_+_87695117 | 2.05 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr2_-_53081199 | 2.01 |
ENSMUST00000239398.2
ENSMUST00000076313.14 ENSMUST00000210789.3 |
Prpf40a
|
pre-mRNA processing factor 40A |
| chr15_-_73056713 | 1.96 |
ENSMUST00000044113.12
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr6_-_148847854 | 1.89 |
ENSMUST00000139355.8
ENSMUST00000146457.2 ENSMUST00000054080.15 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr11_+_33913013 | 1.79 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr10_+_39608094 | 1.78 |
ENSMUST00000019986.13
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr9_-_57552844 | 1.76 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr4_-_3938352 | 1.72 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr10_-_118705029 | 1.68 |
ENSMUST00000004281.10
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr19_+_43678109 | 1.66 |
ENSMUST00000081079.6
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr10_+_88721854 | 1.57 |
ENSMUST00000020255.8
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chr8_-_116434517 | 1.54 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr9_-_75466885 | 1.53 |
ENSMUST00000072232.9
|
Tmod3
|
tropomodulin 3 |
| chr4_+_99952988 | 1.52 |
ENSMUST00000039630.6
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr6_-_38853097 | 1.48 |
ENSMUST00000161779.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr4_-_19708910 | 1.47 |
ENSMUST00000108246.9
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr9_+_108569315 | 1.36 |
ENSMUST00000035220.12
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr4_+_59189239 | 1.30 |
ENSMUST00000030074.8
|
Ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr15_+_102314809 | 1.28 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chr15_+_58287305 | 1.27 |
ENSMUST00000037270.5
|
Fam91a1
|
family with sequence similarity 91, member A1 |
| chr7_-_70010341 | 1.26 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr7_-_16761790 | 1.24 |
ENSMUST00000003183.12
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr11_+_95010935 | 1.22 |
ENSMUST00000092768.7
|
Dlx3
|
distal-less homeobox 3 |
| chr4_-_155306992 | 1.21 |
ENSMUST00000084103.10
ENSMUST00000030917.6 |
Ski
|
ski sarcoma viral oncogene homolog (avian) |
| chr10_+_11157047 | 1.16 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr4_+_135899678 | 1.15 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr10_+_21870565 | 1.12 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_121608859 | 1.06 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr16_-_30207348 | 1.03 |
ENSMUST00000061350.13
ENSMUST00000100013.9 |
Atp13a3
|
ATPase type 13A3 |
| chr10_-_120312374 | 1.02 |
ENSMUST00000072777.14
ENSMUST00000159699.2 |
Hmga2
|
high mobility group AT-hook 2 |
| chr9_+_121946321 | 1.00 |
ENSMUST00000119215.9
ENSMUST00000146832.8 ENSMUST00000118886.9 ENSMUST00000120173.9 ENSMUST00000139181.2 |
Snrk
|
SNF related kinase |
| chr8_+_77626400 | 1.00 |
ENSMUST00000109913.9
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr11_+_28803188 | 0.96 |
ENSMUST00000020759.12
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr4_-_131939548 | 0.95 |
ENSMUST00000152796.8
|
Ythdf2
|
YTH N6-methyladenosine RNA binding protein 2 |
| chr3_-_27764571 | 0.93 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr19_-_31742427 | 0.86 |
ENSMUST00000065067.14
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr15_+_6737853 | 0.86 |
ENSMUST00000061656.8
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr19_-_29783389 | 0.85 |
ENSMUST00000177155.8
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr9_-_71499628 | 0.81 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr9_+_72439496 | 0.81 |
ENSMUST00000163401.9
ENSMUST00000093820.10 |
Rfx7
|
regulatory factor X, 7 |
| chr7_+_110226857 | 0.75 |
ENSMUST00000033054.10
|
Adm
|
adrenomedullin |
| chr13_+_45543208 | 0.74 |
ENSMUST00000038275.11
|
Mylip
|
myosin regulatory light chain interacting protein |
| chr15_-_98505508 | 0.71 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr19_+_32734884 | 0.69 |
ENSMUST00000013807.8
|
Pten
|
phosphatase and tensin homolog |
| chr6_+_115908709 | 0.69 |
ENSMUST00000032471.9
|
Rho
|
rhodopsin |
| chr11_+_20493306 | 0.68 |
ENSMUST00000093292.11
|
Sertad2
|
SERTA domain containing 2 |
| chr7_-_115637970 | 0.66 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr19_+_6356486 | 0.65 |
ENSMUST00000025681.8
|
Cdc42bpg
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr13_-_34529157 | 0.65 |
ENSMUST00000040336.12
|
Slc22a23
|
solute carrier family 22, member 23 |
| chr13_-_64422693 | 0.63 |
ENSMUST00000109770.2
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr19_-_6899173 | 0.61 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr4_-_82423985 | 0.61 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr18_-_39623698 | 0.59 |
ENSMUST00000115567.8
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr1_-_170978157 | 0.58 |
ENSMUST00000155798.2
ENSMUST00000081560.5 ENSMUST00000111336.10 |
Sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr5_+_110283689 | 0.58 |
ENSMUST00000200038.2
ENSMUST00000112519.9 ENSMUST00000198633.5 ENSMUST00000014812.13 ENSMUST00000199811.3 |
Chfr
|
checkpoint with forkhead and ring finger domains |
| chr5_-_96309849 | 0.58 |
ENSMUST00000155901.8
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr19_-_47452840 | 0.56 |
ENSMUST00000081619.10
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr8_-_78451055 | 0.55 |
ENSMUST00000034029.8
|
Ednra
|
endothelin receptor type A |
| chr1_-_57011595 | 0.54 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr7_-_46658144 | 0.53 |
ENSMUST00000061639.10
|
Spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr3_+_88439616 | 0.51 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr6_-_114898739 | 0.51 |
ENSMUST00000032459.14
|
Vgll4
|
vestigial like family member 4 |
| chr9_+_113760002 | 0.48 |
ENSMUST00000084885.12
ENSMUST00000009885.14 |
Ubp1
|
upstream binding protein 1 |
| chr1_+_9868332 | 0.47 |
ENSMUST00000166384.8
ENSMUST00000168907.8 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr5_-_120726721 | 0.46 |
ENSMUST00000046426.10
|
Tpcn1
|
two pore channel 1 |
| chr1_-_134006847 | 0.46 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr17_-_80787398 | 0.46 |
ENSMUST00000068714.7
|
Sos1
|
SOS Ras/Rac guanine nucleotide exchange factor 1 |
| chr14_+_117163215 | 0.45 |
ENSMUST00000078849.12
|
Gpc6
|
glypican 6 |
| chr19_-_5156995 | 0.43 |
ENSMUST00000237438.2
ENSMUST00000236149.2 ENSMUST00000025804.7 |
Rab1b
|
RAB1B, member RAS oncogene family |
| chr15_-_93173032 | 0.43 |
ENSMUST00000057896.5
ENSMUST00000049484.13 ENSMUST00000230063.2 |
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr8_+_89247976 | 0.42 |
ENSMUST00000034086.13
|
Nkd1
|
naked cuticle 1 |
| chr4_+_41465134 | 0.41 |
ENSMUST00000030154.7
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr12_-_73093953 | 0.39 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr11_-_104333059 | 0.34 |
ENSMUST00000106977.8
ENSMUST00000106972.8 |
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr14_-_31552608 | 0.33 |
ENSMUST00000014640.9
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr2_+_18681812 | 0.30 |
ENSMUST00000028071.13
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr16_+_33201164 | 0.29 |
ENSMUST00000165418.9
|
Zfp148
|
zinc finger protein 148 |
| chr6_-_22356098 | 0.29 |
ENSMUST00000165576.8
|
Fam3c
|
family with sequence similarity 3, member C |
| chr12_-_73160181 | 0.29 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr2_+_173501553 | 0.28 |
ENSMUST00000029024.10
ENSMUST00000109110.4 |
Rab22a
|
RAB22A, member RAS oncogene family |
| chr6_-_39702381 | 0.28 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene |
| chr1_-_105284383 | 0.27 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr10_-_91007387 | 0.27 |
ENSMUST00000099355.12
ENSMUST00000105293.11 ENSMUST00000092219.14 ENSMUST00000020123.7 ENSMUST00000072239.14 |
Tmpo
|
thymopoietin |
| chr1_+_131838294 | 0.27 |
ENSMUST00000062264.8
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr9_+_108782646 | 0.27 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr9_+_106048116 | 0.26 |
ENSMUST00000020490.13
|
Wdr82
|
WD repeat domain containing 82 |
| chr6_-_88423464 | 0.25 |
ENSMUST00000204459.3
ENSMUST00000203213.3 ENSMUST00000205179.3 ENSMUST00000165242.4 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr2_+_109721189 | 0.24 |
ENSMUST00000028583.8
|
Lin7c
|
lin-7 homolog C (C. elegans) |
| chr19_-_46033353 | 0.24 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr17_+_28128307 | 0.22 |
ENSMUST00000233525.2
ENSMUST00000025058.15 ENSMUST00000233866.2 |
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr11_+_77576981 | 0.22 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr12_-_56581823 | 0.21 |
ENSMUST00000178477.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr5_-_90514360 | 0.20 |
ENSMUST00000168058.7
ENSMUST00000014421.15 |
Ankrd17
|
ankyrin repeat domain 17 |
| chr11_+_77873535 | 0.19 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr10_-_128481833 | 0.19 |
ENSMUST00000133342.10
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr10_-_44334711 | 0.18 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr8_+_26092252 | 0.16 |
ENSMUST00000136107.9
ENSMUST00000146919.8 ENSMUST00000139966.8 ENSMUST00000142395.8 ENSMUST00000143445.2 |
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr2_+_48704121 | 0.16 |
ENSMUST00000063886.4
|
Acvr2a
|
activin receptor IIA |
| chr2_-_12306722 | 0.15 |
ENSMUST00000028106.11
|
Itga8
|
integrin alpha 8 |
| chr14_-_47805861 | 0.15 |
ENSMUST00000228784.2
ENSMUST00000042988.7 |
Atg14
|
autophagy related 14 |
| chr10_-_121422635 | 0.14 |
ENSMUST00000219493.2
ENSMUST00000020316.4 |
Tbk1
|
TANK-binding kinase 1 |
| chr6_+_21949570 | 0.13 |
ENSMUST00000031680.10
ENSMUST00000115389.8 ENSMUST00000151473.8 |
Ing3
|
inhibitor of growth family, member 3 |
| chr10_+_53473032 | 0.13 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr10_-_56104732 | 0.12 |
ENSMUST00000099739.5
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr6_-_138020409 | 0.11 |
ENSMUST00000111873.8
ENSMUST00000141280.3 |
Slc15a5
|
solute carrier family 15, member 5 |
| chr8_+_91555449 | 0.10 |
ENSMUST00000109614.9
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_74545203 | 0.09 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr16_-_11021116 | 0.09 |
ENSMUST00000230002.2
ENSMUST00000119953.2 |
Rsl1d1
|
ribosomal L1 domain containing 1 |
| chr4_-_129636073 | 0.08 |
ENSMUST00000066257.6
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr4_+_63462984 | 0.07 |
ENSMUST00000035301.7
|
Atp6v1g1
|
ATPase, H+ transporting, lysosomal V1 subunit G1 |
| chr15_+_58805605 | 0.05 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr11_+_87938519 | 0.05 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr19_+_53298906 | 0.04 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chrX_-_102865546 | 0.03 |
ENSMUST00000042664.10
|
Slc16a2
|
solute carrier family 16 (monocarboxylic acid transporters), member 2 |
| chr4_+_126915104 | 0.02 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr2_-_48839276 | 0.02 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr8_+_105067159 | 0.02 |
ENSMUST00000212948.2
ENSMUST00000034343.5 |
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr8_+_13389656 | 0.01 |
ENSMUST00000210165.2
ENSMUST00000170909.2 |
Tfdp1
|
transcription factor Dp 1 |
| chr10_+_86136236 | 0.01 |
ENSMUST00000020234.14
|
Timp3
|
tissue inhibitor of metalloproteinase 3 |
| chr15_-_98769056 | 0.01 |
ENSMUST00000178486.9
ENSMUST00000023741.16 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0060715 | Spemann organizer formation(GO:0060061) syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.8 | 3.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 | 2.8 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.6 | 1.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.6 | 1.8 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.5 | 2.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 1.3 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.4 | 2.5 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.4 | 1.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.3 | 1.2 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.3 | 2.1 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.3 | 1.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.2 | 0.7 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.2 | 0.7 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 0.9 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.2 | 1.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 2.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 0.9 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.2 | 0.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 1.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 1.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 1.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 1.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 1.3 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.9 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.7 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 1.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:2000011 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.1 | 1.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.1 | 0.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.0 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 2.1 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.2 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 1.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.0 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 1.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.6 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) |
| 0.0 | 2.1 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 1.5 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.5 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 2.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.4 | GO:1903020 | positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.6 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 2.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.6 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 0.6 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 2.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 3.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.2 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.1 | 0.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.7 | 2.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.4 | 3.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 2.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 1.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.6 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 3.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.6 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.0 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.6 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.1 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 1.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0034711 | activin-activated receptor activity(GO:0017002) inhibin binding(GO:0034711) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 12.7 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 3.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 3.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 2.0 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 1.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 3.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.7 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |