Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AGAUCAG
Z-value: 0.09
miRNA associated with seed AGAUCAG
| Name | miRBASE accession |
|---|---|
|
mmu-miR-383-5p.2
|
Activity profile of AGAUCAG motif
Sorted Z-values of AGAUCAG motif
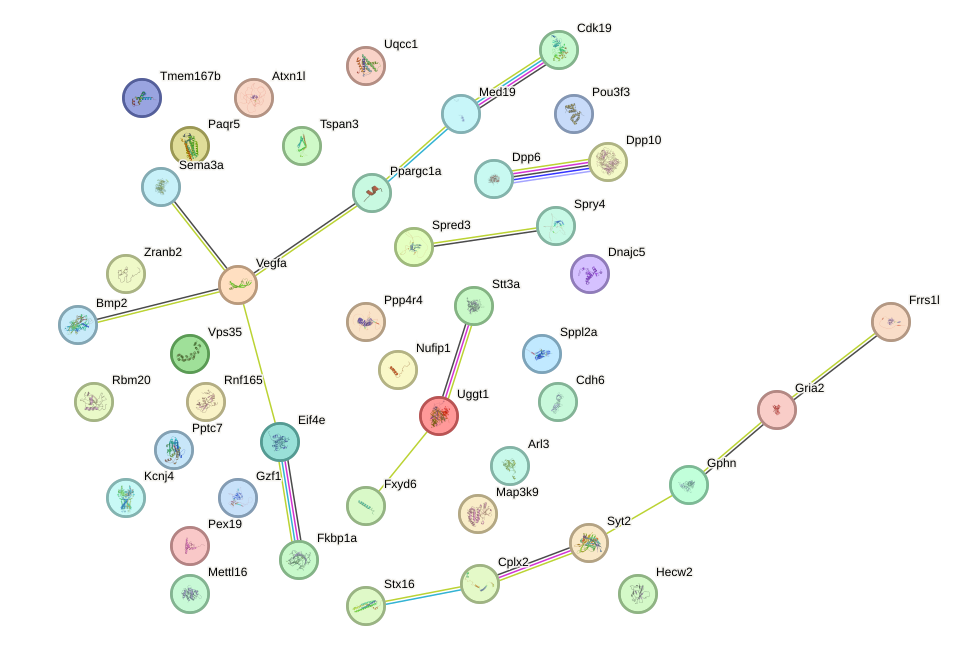
Network of associatons between targets according to the STRING database.
First level regulatory network of AGAUCAG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_6166982 | 0.18 |
ENSMUST00000056416.9
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_-_80710097 | 0.16 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr15_-_79389442 | 0.15 |
ENSMUST00000057801.8
|
Kcnj4
|
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chr4_-_56990306 | 0.14 |
ENSMUST00000053681.6
|
Frrs1l
|
ferric-chelate reductase 1 like |
| chr2_+_96148418 | 0.12 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr13_+_54519161 | 0.11 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr5_+_27109679 | 0.10 |
ENSMUST00000120555.8
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr9_+_45281483 | 0.10 |
ENSMUST00000085939.8
ENSMUST00000217381.2 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr15_-_13173736 | 0.09 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr1_-_123973223 | 0.08 |
ENSMUST00000112606.8
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr1_+_42734889 | 0.08 |
ENSMUST00000054883.4
|
Pou3f3
|
POU domain, class 3, transcription factor 3 |
| chr12_-_81827893 | 0.07 |
ENSMUST00000035987.9
|
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr9_-_56068282 | 0.07 |
ENSMUST00000034876.10
|
Tspan3
|
tetraspanin 3 |
| chr7_-_28868072 | 0.06 |
ENSMUST00000048923.7
|
Spred3
|
sprouty-related EVH1 domain containing 3 |
| chr1_+_134637031 | 0.06 |
ENSMUST00000121990.2
|
Syt2
|
synaptotagmin II |
| chr1_-_54233207 | 0.06 |
ENSMUST00000120904.8
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_-_80411578 | 0.06 |
ENSMUST00000028386.12
|
Nckap1
|
NCK-associated protein 1 |
| chr1_+_171954316 | 0.05 |
ENSMUST00000075895.9
ENSMUST00000111252.4 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr12_+_103498542 | 0.05 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr5_-_51711237 | 0.04 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr1_-_16727067 | 0.04 |
ENSMUST00000188641.7
|
Eloc
|
elongin C |
| chr12_+_78273356 | 0.04 |
ENSMUST00000110388.10
|
Gphn
|
gephyrin |
| chr2_-_155771938 | 0.03 |
ENSMUST00000152766.8
ENSMUST00000139232.8 ENSMUST00000109632.8 ENSMUST00000006036.13 ENSMUST00000142655.2 ENSMUST00000159238.2 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr12_-_14202041 | 0.03 |
ENSMUST00000020926.8
|
Lratd1
|
LRAT domain containing 1 |
| chr18_-_77652820 | 0.03 |
ENSMUST00000026494.14
ENSMUST00000182024.2 |
Rnf165
|
ring finger protein 165 |
| chr18_-_38734389 | 0.03 |
ENSMUST00000025295.8
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr19_-_46561532 | 0.02 |
ENSMUST00000026009.10
ENSMUST00000236255.2 |
Arl3
|
ADP-ribosylation factor-like 3 |
| chr2_+_148522943 | 0.02 |
ENSMUST00000028928.8
|
Gzf1
|
GDNF-inducible zinc finger protein 1 |
| chr2_+_181162278 | 0.02 |
ENSMUST00000072334.12
|
Dnajc5
|
DnaJ heat shock protein family (Hsp40) member C5 |
| chr2_+_133394079 | 0.02 |
ENSMUST00000028836.7
|
Bmp2
|
bone morphogenetic protein 2 |
| chr19_+_53665719 | 0.02 |
ENSMUST00000164202.9
|
Rbm20
|
RNA binding motif protein 20 |
| chr8_-_86026385 | 0.02 |
ENSMUST00000034131.10
|
Vps35
|
VPS35 retromer complex component |
| chr9_-_61934135 | 0.02 |
ENSMUST00000034817.11
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr3_+_138231935 | 0.02 |
ENSMUST00000029803.12
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr11_+_74661647 | 0.02 |
ENSMUST00000010698.13
ENSMUST00000141755.8 |
Mettl16
|
methyltransferase like 16 |
| chr5_-_139761387 | 0.02 |
ENSMUST00000200393.5
ENSMUST00000072607.9 ENSMUST00000196864.2 |
Ints1
|
integrator complex subunit 1 |
| chr2_+_173918715 | 0.02 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr2_+_84508734 | 0.01 |
ENSMUST00000102645.4
|
Med19
|
mediator complex subunit 19 |
| chr14_+_76348312 | 0.01 |
ENSMUST00000022586.2
|
Nufip1
|
nuclear fragile X mental retardation protein interacting protein 1 |
| chr3_-_108469740 | 0.01 |
ENSMUST00000090546.6
|
Tmem167b
|
transmembrane protein 167B |
| chr8_-_110464345 | 0.01 |
ENSMUST00000212605.2
ENSMUST00000093162.4 ENSMUST00000212726.2 |
Atxn1l
|
ataxin 1-like |
| chr2_+_151384403 | 0.01 |
ENSMUST00000044011.12
|
Fkbp1a
|
FK506 binding protein 1a |
| chr2_-_126775136 | 0.01 |
ENSMUST00000028844.11
|
Sppl2a
|
signal peptide peptidase like 2A |
| chr5_+_13449276 | 0.01 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr5_+_122422428 | 0.01 |
ENSMUST00000053426.15
|
Pptc7
|
PTC7 protein phosphatase homolog |
| chr3_+_157239988 | 0.01 |
ENSMUST00000029831.16
ENSMUST00000106057.8 |
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr10_+_40225272 | 0.01 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr17_-_46342739 | 0.01 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr5_+_130248547 | 0.01 |
ENSMUST00000202305.4
ENSMUST00000065329.13 ENSMUST00000200802.4 |
Tmem248
|
transmembrane protein 248 |
| chr9_-_36678868 | 0.01 |
ENSMUST00000217599.2
ENSMUST00000120381.9 |
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr11_-_73671466 | 0.00 |
ENSMUST00000215418.2
ENSMUST00000122224.4 ENSMUST00000124927.3 |
Olfr389
|
olfactory receptor 389 |
| chr1_-_36283326 | 0.00 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.0 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.0 | 0.0 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.0 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |