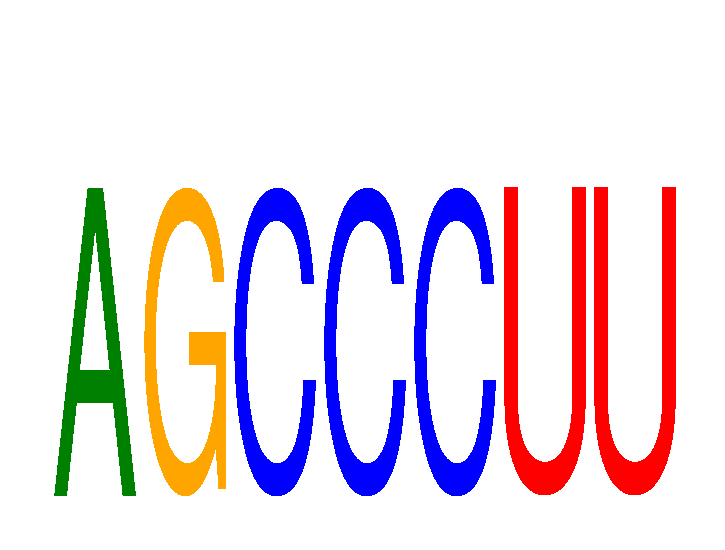Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AGCCCUU
Z-value: 1.75
miRNA associated with seed AGCCCUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-129-1-3p
|
MIMAT0016994 |
|
mmu-miR-129-2-3p
|
MIMAT0000544 |
Activity profile of AGCCCUU motif
Sorted Z-values of AGCCCUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AGCCCUU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_38796539 | 27.13 |
ENSMUST00000163313.3
|
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr4_-_87148672 | 23.69 |
ENSMUST00000107157.9
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr11_-_42073737 | 18.02 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr14_-_55354392 | 15.78 |
ENSMUST00000022819.13
|
Jph4
|
junctophilin 4 |
| chr1_+_152830720 | 15.32 |
ENSMUST00000043313.15
ENSMUST00000186621.2 |
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr1_-_132470672 | 15.04 |
ENSMUST00000086521.11
|
Cntn2
|
contactin 2 |
| chrX_+_165021897 | 14.58 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr17_+_44114894 | 13.42 |
ENSMUST00000044895.13
|
Rcan2
|
regulator of calcineurin 2 |
| chr14_+_111912529 | 13.21 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr7_+_43959637 | 13.17 |
ENSMUST00000107938.8
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr17_+_70276068 | 12.85 |
ENSMUST00000133983.8
|
Dlgap1
|
DLG associated protein 1 |
| chr2_+_83642910 | 12.79 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr8_+_14145848 | 12.64 |
ENSMUST00000152652.8
ENSMUST00000133298.8 |
Dlgap2
|
DLG associated protein 2 |
| chr6_-_37276885 | 12.49 |
ENSMUST00000101532.10
|
Dgki
|
diacylglycerol kinase, iota |
| chr11_-_99134885 | 12.39 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr8_-_9821021 | 12.27 |
ENSMUST00000208933.2
ENSMUST00000110969.5 |
Fam155a
|
family with sequence similarity 155, member A |
| chr5_+_130477642 | 12.26 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr5_+_118307754 | 12.16 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr4_+_120711974 | 11.78 |
ENSMUST00000071093.9
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr2_-_24653059 | 11.74 |
ENSMUST00000100348.10
ENSMUST00000041342.12 ENSMUST00000114447.8 ENSMUST00000102939.9 ENSMUST00000070864.14 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr16_-_9812410 | 11.74 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr4_+_138181616 | 11.58 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr4_-_46991842 | 10.95 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr3_+_7431717 | 10.50 |
ENSMUST00000192468.2
ENSMUST00000028999.12 |
Pkia
|
protein kinase inhibitor, alpha |
| chrX_+_149981074 | 10.45 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr18_-_31580436 | 10.34 |
ENSMUST00000025110.5
|
Syt4
|
synaptotagmin IV |
| chr1_-_154602102 | 10.34 |
ENSMUST00000187541.7
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr11_+_78213791 | 10.30 |
ENSMUST00000017534.15
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr5_-_122917341 | 10.16 |
ENSMUST00000198257.5
ENSMUST00000199599.2 ENSMUST00000196742.2 ENSMUST00000200109.5 ENSMUST00000111668.8 |
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr9_-_21963306 | 9.99 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr7_+_123582021 | 9.91 |
ENSMUST00000106437.2
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr5_+_63806451 | 9.72 |
ENSMUST00000159584.3
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr6_-_136150076 | 9.72 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr17_-_45860580 | 9.57 |
ENSMUST00000180252.3
|
Tmem151b
|
transmembrane protein 151B |
| chr15_+_101122069 | 9.13 |
ENSMUST00000000543.6
|
Tamalin
|
trafficking regulator and scaffold protein tamalin |
| chr5_-_103247920 | 9.05 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr9_+_40180726 | 8.94 |
ENSMUST00000171835.9
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr11_+_104122216 | 8.84 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chr10_+_107107477 | 8.43 |
ENSMUST00000020057.16
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr17_+_24707711 | 8.39 |
ENSMUST00000024958.9
ENSMUST00000234717.2 |
Caskin1
|
CASK interacting protein 1 |
| chr15_-_71599664 | 8.24 |
ENSMUST00000022953.10
|
Fam135b
|
family with sequence similarity 135, member B |
| chr18_-_72484126 | 8.04 |
ENSMUST00000114943.11
|
Dcc
|
deleted in colorectal carcinoma |
| chr7_-_81104423 | 7.77 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr10_-_5872386 | 7.65 |
ENSMUST00000131996.8
ENSMUST00000064225.14 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr1_+_134637031 | 7.47 |
ENSMUST00000121990.2
|
Syt2
|
synaptotagmin II |
| chr4_+_129030710 | 7.45 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr15_+_4404965 | 7.40 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr12_+_95658987 | 7.38 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr2_+_162785394 | 7.10 |
ENSMUST00000035751.12
ENSMUST00000156954.8 |
L3mbtl1
|
L3MBTL1 histone methyl-lysine binding protein |
| chr15_-_78004211 | 7.08 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr7_-_57036920 | 7.07 |
ENSMUST00000068911.13
|
Gabrg3
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 3 |
| chr15_+_80171435 | 7.06 |
ENSMUST00000160424.8
|
Cacna1i
|
calcium channel, voltage-dependent, alpha 1I subunit |
| chr13_-_36918424 | 7.00 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chr11_+_89921121 | 6.99 |
ENSMUST00000092788.4
|
Tmem100
|
transmembrane protein 100 |
| chr7_+_118311740 | 6.94 |
ENSMUST00000106557.8
|
Ccp110
|
centriolar coiled coil protein 110 |
| chr9_-_43027809 | 6.91 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr1_-_22845124 | 6.91 |
ENSMUST00000115273.10
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr11_-_118800314 | 6.81 |
ENSMUST00000117731.8
ENSMUST00000106278.9 ENSMUST00000120061.8 ENSMUST00000017576.11 |
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr5_+_35435663 | 6.78 |
ENSMUST00000049545.7
|
Adra2c
|
adrenergic receptor, alpha 2c |
| chr10_+_3316057 | 6.74 |
ENSMUST00000043374.7
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr8_+_104897074 | 6.68 |
ENSMUST00000164076.3
ENSMUST00000171018.8 ENSMUST00000167633.8 ENSMUST00000093245.13 ENSMUST00000212979.2 |
Bean1
|
brain expressed, associated with Nedd4, 1 |
| chr1_+_166081664 | 6.55 |
ENSMUST00000111416.7
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr1_-_84912810 | 6.49 |
ENSMUST00000027422.7
|
Slc16a14
|
solute carrier family 16 (monocarboxylic acid transporters), member 14 |
| chr13_-_58056089 | 6.48 |
ENSMUST00000185502.7
ENSMUST00000186271.7 ENSMUST00000185905.2 ENSMUST00000187852.7 |
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
| chr17_-_56447332 | 6.45 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr1_+_125604706 | 5.74 |
ENSMUST00000027581.7
|
Gpr39
|
G protein-coupled receptor 39 |
| chr7_+_26958150 | 5.66 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr2_+_121697398 | 5.63 |
ENSMUST00000110586.10
ENSMUST00000078752.10 |
Golm2
|
golgi membrane protein 2 |
| chr18_-_38345010 | 5.60 |
ENSMUST00000159405.3
ENSMUST00000160721.8 |
Pcdh1
|
protocadherin 1 |
| chr11_+_105480796 | 5.48 |
ENSMUST00000168598.8
ENSMUST00000100330.10 |
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr5_+_89034666 | 5.41 |
ENSMUST00000148750.8
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr9_+_58536386 | 5.27 |
ENSMUST00000176250.2
|
Nptn
|
neuroplastin |
| chr3_+_32871669 | 5.24 |
ENSMUST00000072312.12
ENSMUST00000108228.8 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr13_-_43457626 | 5.12 |
ENSMUST00000055341.7
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr19_-_4993060 | 5.10 |
ENSMUST00000133504.2
ENSMUST00000133254.2 ENSMUST00000120475.8 ENSMUST00000025834.15 |
Peli3
|
pellino 3 |
| chr2_-_167032068 | 4.89 |
ENSMUST00000059826.10
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr18_-_62044871 | 4.80 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr2_-_33777874 | 4.74 |
ENSMUST00000041555.10
|
Mvb12b
|
multivesicular body subunit 12B |
| chr4_+_17853452 | 4.61 |
ENSMUST00000029881.10
|
Mmp16
|
matrix metallopeptidase 16 |
| chr12_+_109418759 | 4.57 |
ENSMUST00000056110.15
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr7_-_141925947 | 4.44 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr9_-_108067552 | 4.42 |
ENSMUST00000035208.14
|
Bsn
|
bassoon |
| chr14_+_119092107 | 4.31 |
ENSMUST00000100314.4
|
Cldn10
|
claudin 10 |
| chr3_-_56091096 | 4.26 |
ENSMUST00000029374.8
|
Nbea
|
neurobeachin |
| chr8_-_74080101 | 4.03 |
ENSMUST00000119826.7
ENSMUST00000212459.2 |
Large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr11_-_87878301 | 4.03 |
ENSMUST00000020775.9
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr1_-_170042947 | 3.78 |
ENSMUST00000027979.14
ENSMUST00000123399.2 |
Uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr4_-_35845204 | 3.70 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr11_+_30835358 | 3.68 |
ENSMUST00000109430.2
ENSMUST00000203878.3 |
Gpr75
Asb3
|
G protein-coupled receptor 75 ankyrin repeat and SOCS box-containing 3 |
| chr2_-_52566583 | 3.65 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_+_122988111 | 3.64 |
ENSMUST00000031434.8
ENSMUST00000198602.2 |
Rnf34
|
ring finger protein 34 |
| chrX_+_95757092 | 3.54 |
ENSMUST00000033554.5
|
Gpr165
|
G protein-coupled receptor 165 |
| chr17_+_86475205 | 3.51 |
ENSMUST00000097275.9
|
Prkce
|
protein kinase C, epsilon |
| chr15_-_31367668 | 3.38 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr14_-_33923583 | 3.30 |
ENSMUST00000226613.2
ENSMUST00000096019.4 ENSMUST00000226511.2 |
Gprin2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr14_-_47059694 | 3.18 |
ENSMUST00000111817.8
ENSMUST00000079314.12 |
Gmfb
|
glia maturation factor, beta |
| chrX_+_100892981 | 3.08 |
ENSMUST00000124279.6
ENSMUST00000101339.8 |
Nhsl2
|
NHS-like 2 |
| chr3_-_8732316 | 3.05 |
ENSMUST00000042412.5
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1 |
| chr7_-_45516553 | 3.04 |
ENSMUST00000002848.10
|
Grin2d
|
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
| chr2_+_179684288 | 2.86 |
ENSMUST00000041126.9
|
Ss18l1
|
SS18, nBAF chromatin remodeling complex subunit like 1 |
| chr13_-_41373870 | 2.85 |
ENSMUST00000021793.15
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr7_+_125871761 | 2.84 |
ENSMUST00000056028.11
|
Sbk1
|
SH3-binding kinase 1 |
| chr15_-_79718423 | 2.78 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr12_+_64964674 | 2.77 |
ENSMUST00000058135.6
ENSMUST00000220993.2 |
Gm527
|
predicted gene 527 |
| chr1_-_131066004 | 2.75 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr7_+_48896560 | 2.69 |
ENSMUST00000184945.8
|
Nav2
|
neuron navigator 2 |
| chr16_-_37205277 | 2.63 |
ENSMUST00000114787.8
ENSMUST00000114782.8 ENSMUST00000114775.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr14_+_62793175 | 2.62 |
ENSMUST00000039064.8
|
Fam124a
|
family with sequence similarity 124, member A |
| chr8_+_111760521 | 2.61 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr9_+_45749869 | 2.59 |
ENSMUST00000078111.11
ENSMUST00000034591.11 |
Bace1
|
beta-site APP cleaving enzyme 1 |
| chr4_+_42949814 | 2.57 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr10_-_33972503 | 2.57 |
ENSMUST00000069125.8
|
Calhm5
|
calcium homeostasis modulator family member 5 |
| chr7_+_106969950 | 2.49 |
ENSMUST00000073459.12
|
Syt9
|
synaptotagmin IX |
| chr5_-_73805063 | 2.47 |
ENSMUST00000081170.9
|
Sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr1_+_180396478 | 2.45 |
ENSMUST00000027777.12
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
| chr6_-_142910094 | 2.44 |
ENSMUST00000032421.4
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr7_+_109721230 | 2.42 |
ENSMUST00000033326.10
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr11_-_65679101 | 2.41 |
ENSMUST00000152096.8
ENSMUST00000046963.10 |
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr4_-_45826923 | 2.29 |
ENSMUST00000044297.7
|
Igfbpl1
|
insulin-like growth factor binding protein-like 1 |
| chr2_-_37537224 | 2.28 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr10_+_88215079 | 2.22 |
ENSMUST00000130301.8
ENSMUST00000020251.10 |
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr19_-_4319170 | 2.22 |
ENSMUST00000037992.16
ENSMUST00000113852.6 ENSMUST00000236794.2 |
Ssh3
|
slingshot protein phosphatase 3 |
| chr2_-_6889783 | 2.19 |
ENSMUST00000170438.8
ENSMUST00000114924.10 ENSMUST00000114934.11 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr10_-_94780695 | 2.15 |
ENSMUST00000099337.5
|
Plxnc1
|
plexin C1 |
| chr11_+_3280401 | 2.13 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr4_+_136013372 | 2.12 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr4_+_73931679 | 2.11 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr1_+_155034452 | 2.11 |
ENSMUST00000027743.13
ENSMUST00000195302.6 |
Stx6
|
syntaxin 6 |
| chr15_+_97682210 | 2.10 |
ENSMUST00000117892.2
ENSMUST00000229084.2 |
Slc48a1
|
solute carrier family 48 (heme transporter), member 1 |
| chr2_+_156562989 | 2.10 |
ENSMUST00000000094.14
|
Dlgap4
|
DLG associated protein 4 |
| chr18_-_75830595 | 2.08 |
ENSMUST00000165559.3
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chrX_-_23231245 | 2.01 |
ENSMUST00000115313.8
|
Klhl13
|
kelch-like 13 |
| chr7_+_30193047 | 2.01 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr1_-_157240138 | 1.96 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr5_+_101912939 | 1.84 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr1_+_92759324 | 1.84 |
ENSMUST00000045970.8
|
Gpc1
|
glypican 1 |
| chr17_-_66079681 | 1.84 |
ENSMUST00000070673.9
|
Rab31
|
RAB31, member RAS oncogene family |
| chr4_-_57301791 | 1.67 |
ENSMUST00000075637.11
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr19_+_55304703 | 1.65 |
ENSMUST00000225529.2
ENSMUST00000223690.2 ENSMUST00000095950.3 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr4_+_109137462 | 1.65 |
ENSMUST00000102729.10
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr2_+_158251761 | 1.64 |
ENSMUST00000109486.9
ENSMUST00000046274.12 |
Ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr9_-_117081518 | 1.59 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chrX_-_103714653 | 1.55 |
ENSMUST00000042070.6
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr9_-_71393175 | 1.53 |
ENSMUST00000233263.2
ENSMUST00000034720.12 |
Polr2m
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr18_+_44960813 | 1.50 |
ENSMUST00000037763.11
|
Ythdc2
|
YTH domain containing 2 |
| chrX_+_74557905 | 1.48 |
ENSMUST00000114070.10
ENSMUST00000033540.6 |
Vbp1
|
von Hippel-Lindau binding protein 1 |
| chr5_+_115567644 | 1.45 |
ENSMUST00000150779.8
|
Msi1
|
musashi RNA-binding protein 1 |
| chr6_-_32565127 | 1.43 |
ENSMUST00000115096.4
|
Plxna4
|
plexin A4 |
| chr1_-_192812509 | 1.37 |
ENSMUST00000085555.7
|
Utp25
|
UTP25 small subunit processome component |
| chr11_+_80191692 | 1.33 |
ENSMUST00000017836.8
|
Rhbdl3
|
rhomboid like 3 |
| chr12_+_102724223 | 1.29 |
ENSMUST00000046404.8
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr3_+_125197722 | 1.21 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr15_+_99615396 | 1.20 |
ENSMUST00000023760.13
ENSMUST00000162194.2 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr6_+_36364990 | 1.18 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr1_-_136273811 | 1.16 |
ENSMUST00000048309.12
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr5_+_67417908 | 1.14 |
ENSMUST00000037918.12
ENSMUST00000162543.8 ENSMUST00000161233.8 ENSMUST00000160352.8 |
Tmem33
|
transmembrane protein 33 |
| chr6_+_115830431 | 1.14 |
ENSMUST00000112925.8
ENSMUST00000038234.13 ENSMUST00000112923.7 |
Ift122
|
intraflagellar transport 122 |
| chr13_+_120151982 | 1.10 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr19_-_7319157 | 1.09 |
ENSMUST00000164205.8
ENSMUST00000165286.8 ENSMUST00000168324.2 ENSMUST00000032557.15 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr16_+_72460029 | 1.08 |
ENSMUST00000023600.8
|
Robo1
|
roundabout guidance receptor 1 |
| chr10_-_110845860 | 1.04 |
ENSMUST00000041723.15
|
Zdhhc17
|
zinc finger, DHHC domain containing 17 |
| chr2_+_157266175 | 1.02 |
ENSMUST00000029175.14
ENSMUST00000092576.11 |
Src
|
Rous sarcoma oncogene |
| chr9_-_96634874 | 1.02 |
ENSMUST00000152594.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr6_+_113355076 | 1.00 |
ENSMUST00000156898.5
ENSMUST00000203578.3 ENSMUST00000171058.8 |
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr2_+_20742115 | 0.99 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr11_-_115918784 | 0.98 |
ENSMUST00000106454.8
|
H3f3b
|
H3.3 histone B |
| chr18_-_38734389 | 0.94 |
ENSMUST00000025295.8
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr1_+_152275575 | 0.92 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr2_-_25982160 | 0.91 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr7_-_118183892 | 0.90 |
ENSMUST00000044195.6
|
Tmc7
|
transmembrane channel-like gene family 7 |
| chr7_-_27990555 | 0.88 |
ENSMUST00000056589.15
|
Selenov
|
selenoprotein V |
| chr1_+_184936306 | 0.88 |
ENSMUST00000194740.6
ENSMUST00000069652.8 |
Rab3gap2
|
RAB3 GTPase activating protein subunit 2 |
| chr13_-_30170031 | 0.88 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr2_-_124965537 | 0.88 |
ENSMUST00000142718.8
ENSMUST00000152367.8 ENSMUST00000067780.10 ENSMUST00000147105.8 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr11_-_85030761 | 0.87 |
ENSMUST00000108075.9
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr7_-_12552764 | 0.84 |
ENSMUST00000108546.2
ENSMUST00000072222.8 |
Zfp329
|
zinc finger protein 329 |
| chr5_-_149559667 | 0.82 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr2_+_163916042 | 0.82 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr9_-_114762986 | 0.81 |
ENSMUST00000146623.8
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr9_-_63665216 | 0.81 |
ENSMUST00000034973.10
|
Smad3
|
SMAD family member 3 |
| chr16_+_21828223 | 0.80 |
ENSMUST00000023561.8
|
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr1_+_191449946 | 0.76 |
ENSMUST00000133076.7
ENSMUST00000110855.8 |
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr11_+_110289941 | 0.74 |
ENSMUST00000020949.12
ENSMUST00000100260.2 |
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr11_+_5905693 | 0.74 |
ENSMUST00000002818.9
|
Ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr14_+_20724366 | 0.73 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr1_+_59802543 | 0.67 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr4_+_105014536 | 0.66 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr2_+_52747855 | 0.65 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr7_+_63937413 | 0.64 |
ENSMUST00000032736.11
|
Mtmr10
|
myotubularin related protein 10 |
| chr11_-_106107132 | 0.64 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr14_-_30075424 | 0.58 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr12_-_65010124 | 0.57 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr4_+_21848039 | 0.56 |
ENSMUST00000098238.9
ENSMUST00000108229.2 |
Pnisr
|
PNN interacting serine/arginine-rich |
| chr4_+_148225139 | 0.56 |
ENSMUST00000140049.8
ENSMUST00000105707.2 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr1_+_171156568 | 0.47 |
ENSMUST00000111300.8
|
Dedd
|
death effector domain-containing |
| chr2_-_73143045 | 0.39 |
ENSMUST00000058615.10
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr17_+_29768757 | 0.36 |
ENSMUST00000048677.9
ENSMUST00000150388.3 |
Tbc1d22b
Gm28052
|
TBC1 domain family, member 22B predicted gene, 28052 |
| chr11_-_5331734 | 0.29 |
ENSMUST00000172492.8
|
Znrf3
|
zinc and ring finger 3 |
| chr1_+_64571942 | 0.19 |
ENSMUST00000171164.8
ENSMUST00000187811.7 ENSMUST00000049932.12 ENSMUST00000087366.11 |
Creb1
|
cAMP responsive element binding protein 1 |
| chr4_-_62278673 | 0.16 |
ENSMUST00000084527.10
ENSMUST00000098033.10 |
Fkbp15
|
FK506 binding protein 15 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 3.5 | 10.5 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 3.4 | 10.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 2.6 | 25.7 | GO:0046959 | habituation(GO:0046959) |
| 2.6 | 15.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 2.0 | 12.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 1.9 | 9.7 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 1.8 | 14.6 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 8.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 1.5 | 4.4 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.5 | 11.7 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.4 | 8.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.4 | 6.9 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.3 | 5.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.3 | 5.2 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 1.3 | 9.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.2 | 1.2 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.2 | 6.9 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 1.1 | 10.3 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.1 | 7.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 1.0 | 5.1 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.0 | 3.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 1.0 | 7.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.0 | 12.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.0 | 6.8 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.0 | 5.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.9 | 8.8 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.9 | 2.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.9 | 10.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.8 | 12.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.8 | 2.5 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.8 | 4.9 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.8 | 9.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.8 | 15.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.8 | 18.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.8 | 7.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.7 | 2.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.7 | 7.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.7 | 3.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.7 | 2.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.7 | 2.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 1.8 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.6 | 3.6 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.6 | 4.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.6 | 10.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 13.2 | GO:0021756 | striatum development(GO:0021756) |
| 0.5 | 2.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.5 | 1.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.5 | 11.7 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.5 | 10.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 18.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 13.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.5 | 21.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 23.7 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.5 | 7.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.5 | 3.2 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.4 | 5.7 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.4 | 1.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.4 | 1.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 1.5 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.4 | 4.0 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 1.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 4.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 1.5 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 0.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.3 | 2.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 7.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.3 | 1.6 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.3 | 4.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 2.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 2.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 7.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.2 | 0.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 5.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.0 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 0.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 2.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 2.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.2 | 3.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 5.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.6 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 1.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.1 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 4.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 3.0 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 3.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.5 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 0.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.8 | GO:0051135 | positive regulation of NK T cell activation(GO:0051135) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 1.6 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 2.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 12.6 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.1 | 3.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.8 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 3.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.7 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.1 | 0.2 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 | 7.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 2.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 2.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 2.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 4.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.9 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.6 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 4.6 | GO:0048524 | positive regulation of viral process(GO:0048524) |
| 0.0 | 4.3 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 2.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 2.6 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.7 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 6.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.0 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 2.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.8 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 5.2 | GO:1990138 | neuron projection extension(GO:1990138) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 5.3 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.9 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 1.1 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 2.3 | GO:0006457 | protein folding(GO:0006457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.9 | 11.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.5 | 37.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.4 | 15.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.1 | 8.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.0 | 8.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.6 | 7.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.6 | 18.0 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.6 | 12.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 12.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.6 | 8.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.6 | 8.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 3.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.5 | 33.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.5 | 21.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.4 | 16.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 2.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 4.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 11.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 2.5 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.2 | 5.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 4.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 81.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 25.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 1.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 7.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 2.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 2.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 18.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 6.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 6.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 62.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 13.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 5.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 8.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 3.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 5.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 3.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 5.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 7.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 2.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 56.2 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.2 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 3.9 | 23.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 3.4 | 10.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 2.6 | 15.3 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 2.5 | 27.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) |
| 2.3 | 11.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.9 | 13.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.8 | 10.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.8 | 9.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.8 | 12.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.8 | 7.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 1.8 | 8.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.8 | 7.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.7 | 6.8 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.6 | 14.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.5 | 13.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.2 | 7.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 1.2 | 8.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.1 | 8.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.0 | 5.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.0 | 10.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 26.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.0 | 8.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.9 | 3.5 | GO:0035276 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.9 | 2.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.8 | 6.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.7 | 10.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.6 | 1.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.6 | 2.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.5 | 2.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 7.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 1.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 10.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 2.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 2.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.4 | 3.4 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.4 | 10.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.4 | 1.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.4 | 2.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.4 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 6.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 6.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 6.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 3.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.3 | 1.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 7.1 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.2 | 5.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 2.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 2.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 4.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 3.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 10.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 0.8 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 3.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 7.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 4.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 4.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.8 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 4.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 15.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 4.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 6.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 2.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.7 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 4.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 14.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 5.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 7.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 8.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 5.4 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 10.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 3.6 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 14.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 12.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 12.5 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 5.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 23.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.6 | 9.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 12.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 18.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 2.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 12.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 11.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 7.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 3.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 6.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 6.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 11.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 6.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 25.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 24.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.6 | 9.6 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.6 | 8.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.5 | 10.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 4.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.5 | 6.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 16.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 11.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.4 | 25.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 13.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 8.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 5.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 10.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 29.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 8.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 7.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 13.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 4.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 4.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 4.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 11.7 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 4.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 4.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 4.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |