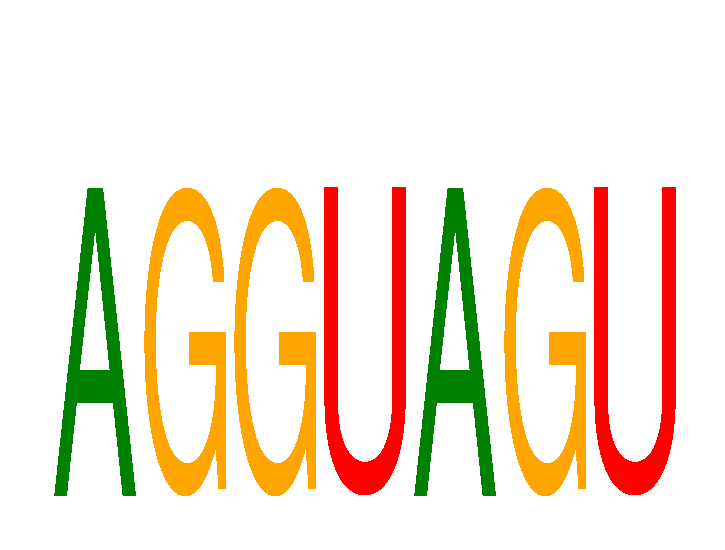Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AGGUAGU
Z-value: 0.53
miRNA associated with seed AGGUAGU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-196a-5p
|
MIMAT0000518 |
|
mmu-miR-196b-5p
|
MIMAT0001081 |
Activity profile of AGGUAGU motif
Sorted Z-values of AGGUAGU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AGGUAGU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_45350698 | 3.31 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr9_-_75518585 | 3.05 |
ENSMUST00000098552.10
ENSMUST00000064433.11 |
Tmod2
|
tropomodulin 2 |
| chr9_+_111140741 | 2.98 |
ENSMUST00000078626.8
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr11_+_94827050 | 2.67 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr1_+_86230931 | 2.44 |
ENSMUST00000113306.4
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr9_+_65008735 | 2.39 |
ENSMUST00000213533.2
ENSMUST00000035499.5 ENSMUST00000077696.13 ENSMUST00000166273.2 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr6_+_4505493 | 2.19 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr1_-_168259465 | 2.11 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chrX_+_35592006 | 1.93 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr17_-_68311073 | 1.85 |
ENSMUST00000024840.12
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr11_+_67477347 | 1.77 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr7_-_16657825 | 1.73 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3 |
| chr11_-_76737794 | 1.60 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr1_-_37580084 | 1.52 |
ENSMUST00000151952.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr4_+_54947976 | 1.47 |
ENSMUST00000098070.10
|
Zfp462
|
zinc finger protein 462 |
| chr1_+_20960819 | 1.45 |
ENSMUST00000189400.7
|
Paqr8
|
progestin and adipoQ receptor family member VIII |
| chr3_+_68375495 | 1.41 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr18_-_15536747 | 1.41 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr10_+_117465397 | 1.38 |
ENSMUST00000020399.6
|
Cpm
|
carboxypeptidase M |
| chr19_+_23736205 | 1.34 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr12_+_100165694 | 1.32 |
ENSMUST00000110082.11
|
Calm1
|
calmodulin 1 |
| chr2_-_93164812 | 1.26 |
ENSMUST00000111265.9
|
Tspan18
|
tetraspanin 18 |
| chr1_-_135032972 | 1.21 |
ENSMUST00000044828.14
|
Lgr6
|
leucine-rich repeat-containing G protein-coupled receptor 6 |
| chr3_-_132940647 | 1.20 |
ENSMUST00000147041.10
ENSMUST00000161022.9 |
Arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr15_+_102898966 | 1.17 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr16_+_32882882 | 1.16 |
ENSMUST00000023497.3
|
Lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr2_+_31135813 | 1.14 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chr11_+_96177449 | 1.08 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr2_-_152256947 | 1.04 |
ENSMUST00000099207.5
|
Zcchc3
|
zinc finger, CCHC domain containing 3 |
| chr7_+_44091822 | 1.01 |
ENSMUST00000058667.15
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr10_-_120312374 | 1.00 |
ENSMUST00000072777.14
ENSMUST00000159699.2 |
Hmga2
|
high mobility group AT-hook 2 |
| chr7_+_15864265 | 1.00 |
ENSMUST00000168693.3
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr5_-_135573962 | 0.99 |
ENSMUST00000060311.12
|
Hip1
|
huntingtin interacting protein 1 |
| chr2_-_34262012 | 0.97 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr7_+_120442048 | 0.97 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr7_+_90075762 | 0.97 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr6_-_144994534 | 0.96 |
ENSMUST00000032402.12
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr15_+_11064887 | 0.95 |
ENSMUST00000061318.9
|
Adamts12
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 12 |
| chr6_+_4747298 | 0.94 |
ENSMUST00000166678.2
ENSMUST00000176204.8 |
Peg10
|
paternally expressed 10 |
| chr17_-_81035453 | 0.93 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr9_+_102988940 | 0.93 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr9_-_107109108 | 0.88 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr13_+_35843816 | 0.84 |
ENSMUST00000075220.14
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr19_+_28941292 | 0.83 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr15_-_43034205 | 0.82 |
ENSMUST00000063492.8
ENSMUST00000226810.2 |
Rspo2
|
R-spondin 2 |
| chr13_-_117162041 | 0.81 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr16_-_97412169 | 0.81 |
ENSMUST00000232141.2
ENSMUST00000000395.8 |
Tmprss2
|
transmembrane protease, serine 2 |
| chr19_-_7360350 | 0.79 |
ENSMUST00000025924.4
|
Spindoc
|
spindlin interactor and repressor of chromatin binding |
| chr6_+_54572096 | 0.75 |
ENSMUST00000119706.8
|
Plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr8_-_91074971 | 0.74 |
ENSMUST00000109621.10
|
Tox3
|
TOX high mobility group box family member 3 |
| chr8_+_117884711 | 0.74 |
ENSMUST00000064488.11
ENSMUST00000162997.3 |
Gan
|
giant axonal neuropathy |
| chr6_-_52203146 | 0.71 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr10_-_76181089 | 0.70 |
ENSMUST00000036033.14
ENSMUST00000160048.8 ENSMUST00000105417.10 |
Dip2a
|
disco interacting protein 2 homolog A |
| chr8_-_47805383 | 0.64 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr8_+_27513819 | 0.64 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr5_-_107437427 | 0.63 |
ENSMUST00000031224.15
|
Tgfbr3
|
transforming growth factor, beta receptor III |
| chr13_-_8921732 | 0.63 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr11_-_50822438 | 0.62 |
ENSMUST00000109124.10
|
Zfp354b
|
zinc finger protein 354B |
| chrX_+_139565657 | 0.59 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr9_-_117081518 | 0.52 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr15_+_87509413 | 0.51 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr3_+_93349637 | 0.50 |
ENSMUST00000064257.6
|
Tchh
|
trichohyalin |
| chr4_+_28813125 | 0.49 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr11_+_96183294 | 0.49 |
ENSMUST00000173432.3
|
Hoxb6
|
homeobox B6 |
| chr12_-_11486544 | 0.48 |
ENSMUST00000072299.7
|
Vsnl1
|
visinin-like 1 |
| chr3_-_10505113 | 0.48 |
ENSMUST00000029047.12
ENSMUST00000195822.2 ENSMUST00000099223.11 |
Snx16
|
sorting nexin 16 |
| chrX_+_92698469 | 0.48 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr18_+_11052458 | 0.47 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr11_+_54194624 | 0.47 |
ENSMUST00000093106.12
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr3_-_104419128 | 0.46 |
ENSMUST00000199070.5
ENSMUST00000046316.11 ENSMUST00000198332.2 |
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr1_-_134883645 | 0.46 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_+_14901343 | 0.44 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chrX_-_103244784 | 0.44 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr16_-_46317318 | 0.43 |
ENSMUST00000023335.13
ENSMUST00000023334.15 |
Nectin3
|
nectin cell adhesion molecule 3 |
| chr15_-_6904450 | 0.43 |
ENSMUST00000022746.13
ENSMUST00000176826.2 |
Osmr
|
oncostatin M receptor |
| chr14_-_70680882 | 0.38 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr8_+_24159669 | 0.36 |
ENSMUST00000042352.11
ENSMUST00000207301.2 |
Zmat4
|
zinc finger, matrin type 4 |
| chr1_+_59802543 | 0.36 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr12_-_78908409 | 0.36 |
ENSMUST00000021536.9
|
Atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chr6_-_52181393 | 0.35 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr1_-_190758321 | 0.35 |
ENSMUST00000191946.2
ENSMUST00000085635.6 |
Flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr6_-_52194440 | 0.33 |
ENSMUST00000048715.9
|
Hoxa7
|
homeobox A7 |
| chr14_-_49304110 | 0.33 |
ENSMUST00000162175.9
|
Exoc5
|
exocyst complex component 5 |
| chr13_+_97061182 | 0.33 |
ENSMUST00000171324.3
|
Gcnt4
|
glucosaminyl (N-acetyl) transferase 4, core 2 (beta-1,6-N-acetylglucosaminyltransferase) |
| chr8_-_27664651 | 0.32 |
ENSMUST00000054212.7
ENSMUST00000033878.14 ENSMUST00000209377.2 |
Rab11fip1
|
RAB11 family interacting protein 1 (class I) |
| chr2_+_28531239 | 0.31 |
ENSMUST00000028155.12
ENSMUST00000113869.8 ENSMUST00000113867.9 |
Tsc1
|
TSC complex subunit 1 |
| chr15_+_10177709 | 0.29 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor |
| chr13_+_45660905 | 0.29 |
ENSMUST00000000260.13
|
Gmpr
|
guanosine monophosphate reductase |
| chr9_+_27702243 | 0.29 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chrX_+_55655111 | 0.26 |
ENSMUST00000144068.8
ENSMUST00000077741.12 ENSMUST00000114784.4 |
Slc9a6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6 |
| chr2_-_152239966 | 0.24 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr5_+_53713137 | 0.24 |
ENSMUST00000087360.9
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_-_153371861 | 0.24 |
ENSMUST00000035346.14
|
Nol4l
|
nucleolar protein 4-like |
| chr4_+_21931290 | 0.24 |
ENSMUST00000029908.8
|
Faxc
|
failed axon connections homolog |
| chr11_+_54329014 | 0.24 |
ENSMUST00000046835.14
|
Fnip1
|
folliculin interacting protein 1 |
| chrX_-_13712746 | 0.22 |
ENSMUST00000115436.9
ENSMUST00000033321.11 ENSMUST00000115438.10 |
Cask
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr11_+_105480796 | 0.22 |
ENSMUST00000168598.8
ENSMUST00000100330.10 |
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr8_-_92039850 | 0.21 |
ENSMUST00000047783.14
|
Rpgrip1l
|
Rpgrip1-like |
| chrX_-_110606766 | 0.21 |
ENSMUST00000113422.9
ENSMUST00000038472.7 |
Hdx
|
highly divergent homeobox |
| chr14_-_105414294 | 0.18 |
ENSMUST00000163545.8
|
Rbm26
|
RNA binding motif protein 26 |
| chr10_-_63926044 | 0.17 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_102965601 | 0.16 |
ENSMUST00000029445.13
ENSMUST00000200069.5 |
Nras
|
neuroblastoma ras oncogene |
| chr11_+_44508137 | 0.15 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr4_+_6365650 | 0.15 |
ENSMUST00000029912.11
ENSMUST00000103008.12 |
Sdcbp
|
syndecan binding protein |
| chr8_-_84831391 | 0.14 |
ENSMUST00000041367.9
ENSMUST00000210279.2 |
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr2_-_37333109 | 0.13 |
ENSMUST00000102789.3
ENSMUST00000067043.5 ENSMUST00000112932.2 |
Zbtb26
Zbtb6
|
zinc finger and BTB domain containing 26 zinc finger and BTB domain containing 6 |
| chr11_-_57723502 | 0.13 |
ENSMUST00000160392.9
ENSMUST00000108845.3 |
Hand1
|
heart and neural crest derivatives expressed 1 |
| chr2_+_156038562 | 0.13 |
ENSMUST00000037401.10
|
Phf20
|
PHD finger protein 20 |
| chr6_-_127128007 | 0.13 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr10_-_5872386 | 0.12 |
ENSMUST00000131996.8
ENSMUST00000064225.14 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr19_+_30007910 | 0.12 |
ENSMUST00000025739.14
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chr6_+_86826470 | 0.11 |
ENSMUST00000089519.13
ENSMUST00000204414.3 |
Aak1
|
AP2 associated kinase 1 |
| chr4_-_82777746 | 0.11 |
ENSMUST00000156055.2
ENSMUST00000030110.15 |
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr3_+_144998233 | 0.11 |
ENSMUST00000029848.5
ENSMUST00000139001.2 |
Col24a1
|
collagen, type XXIV, alpha 1 |
| chr2_+_154278357 | 0.09 |
ENSMUST00000109725.8
ENSMUST00000099178.10 ENSMUST00000045270.15 ENSMUST00000109724.2 |
Cbfa2t2
|
CBFA2/RUNX1 translocation partner 2 |
| chr10_-_61219229 | 0.09 |
ENSMUST00000020289.10
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr18_+_12261798 | 0.08 |
ENSMUST00000025270.8
|
Riok3
|
RIO kinase 3 |
| chr16_-_67417768 | 0.08 |
ENSMUST00000114292.8
ENSMUST00000120898.8 |
Cadm2
|
cell adhesion molecule 2 |
| chr14_-_124914516 | 0.08 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chrX_-_7054952 | 0.07 |
ENSMUST00000004428.14
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr10_-_43416941 | 0.07 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr17_+_34148485 | 0.07 |
ENSMUST00000047503.16
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr6_+_92068361 | 0.06 |
ENSMUST00000113460.8
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_+_121358778 | 0.05 |
ENSMUST00000036025.16
ENSMUST00000112621.2 |
Ccdc93
|
coiled-coil domain containing 93 |
| chr10_+_127512933 | 0.05 |
ENSMUST00000118612.8
ENSMUST00000048099.5 |
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr12_+_117480099 | 0.05 |
ENSMUST00000109691.4
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr10_-_23225781 | 0.05 |
ENSMUST00000092665.12
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr5_+_13449276 | 0.04 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr16_+_37736776 | 0.04 |
ENSMUST00000061274.9
|
Gpr156
|
G protein-coupled receptor 156 |
| chr5_-_124233812 | 0.03 |
ENSMUST00000031354.11
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr12_-_91556761 | 0.02 |
ENSMUST00000021345.14
|
Gtf2a1
|
general transcription factor II A, 1 |
| chrX_-_20928219 | 0.02 |
ENSMUST00000040628.6
ENSMUST00000115333.9 ENSMUST00000115334.8 |
Zfp182
|
zinc finger protein 182 |
| chrX_+_152020744 | 0.01 |
ENSMUST00000112574.9
|
Klf8
|
Kruppel-like factor 8 |
| chr13_+_25240138 | 0.01 |
ENSMUST00000069614.7
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr11_-_115494692 | 0.01 |
ENSMUST00000125097.2
ENSMUST00000019135.14 ENSMUST00000106508.10 |
Gga3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chrX_-_160942713 | 0.01 |
ENSMUST00000087085.10
|
Nhs
|
NHS actin remodeling regulator |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 | 1.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 2.7 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 1.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.3 | 0.8 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 1.0 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.2 | 0.9 | GO:1902202 | proteoglycan catabolic process(GO:0030167) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 3.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.2 | 0.6 | GO:0060935 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.2 | 0.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 2.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 1.0 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.4 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.3 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.5 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.1 | 0.5 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 1.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.3 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 2.0 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 1.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 1.0 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 1.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 1.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.4 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 1.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 3.3 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.3 | 3.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 3.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 3.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.5 | 8.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 1.0 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.2 | 3.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.8 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 1.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 3.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.0 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 1.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.4 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 2.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.5 | GO:0008046 | GPI-linked ephrin receptor activity(GO:0005004) axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 3.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 3.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |