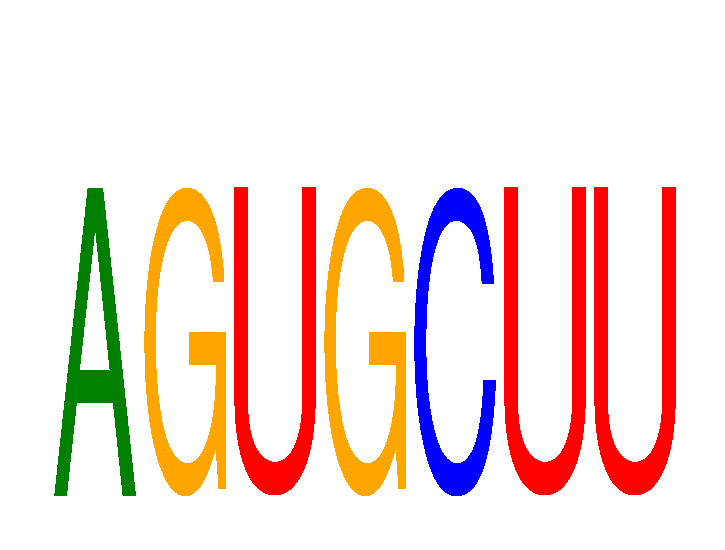Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AGUGCUU
Z-value: 0.66
miRNA associated with seed AGUGCUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-302c-3p
|
MIMAT0003376 |
Activity profile of AGUGCUU motif
Sorted Z-values of AGUGCUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AGUGCUU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_55033113 | 5.29 |
ENSMUST00000038926.13
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr11_-_117859997 | 4.40 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr17_-_32074754 | 4.11 |
ENSMUST00000024839.6
|
Sik1
|
salt inducible kinase 1 |
| chr5_-_136596299 | 4.03 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chr2_+_119156265 | 3.78 |
ENSMUST00000102517.4
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr19_+_41471067 | 3.76 |
ENSMUST00000067795.13
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr8_+_84682136 | 3.65 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr16_+_42727926 | 3.33 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr5_-_21156766 | 3.28 |
ENSMUST00000036489.10
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr1_-_13660027 | 3.25 |
ENSMUST00000027068.11
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr9_+_72439496 | 3.18 |
ENSMUST00000163401.9
ENSMUST00000093820.10 |
Rfx7
|
regulatory factor X, 7 |
| chr2_-_104647041 | 3.06 |
ENSMUST00000117237.2
ENSMUST00000231375.2 |
Qser1
|
glutamine and serine rich 1 |
| chr9_+_30941924 | 2.93 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr15_+_102314809 | 2.92 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chr11_-_117671436 | 2.87 |
ENSMUST00000026659.10
ENSMUST00000127227.2 |
Tmc6
|
transmembrane channel-like gene family 6 |
| chr1_-_69724939 | 2.82 |
ENSMUST00000027146.9
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr11_-_103158190 | 2.79 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr18_+_84106188 | 2.75 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr12_-_79343040 | 2.69 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr6_+_38410848 | 2.66 |
ENSMUST00000160583.8
|
Ubn2
|
ubinuclein 2 |
| chr8_-_41586713 | 2.62 |
ENSMUST00000155055.2
ENSMUST00000059115.13 ENSMUST00000145860.2 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr7_-_115637970 | 2.57 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr18_-_10182007 | 2.50 |
ENSMUST00000067947.7
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr10_-_12839995 | 2.45 |
ENSMUST00000219727.2
ENSMUST00000163425.9 ENSMUST00000042861.7 ENSMUST00000218685.2 |
Stx11
|
syntaxin 11 |
| chr16_+_65612083 | 2.40 |
ENSMUST00000168064.3
|
Vgll3
|
vestigial like family member 3 |
| chr1_-_82269137 | 2.38 |
ENSMUST00000069799.3
|
Irs1
|
insulin receptor substrate 1 |
| chr14_+_32321341 | 2.38 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr10_+_70080913 | 2.35 |
ENSMUST00000046807.7
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chr12_+_71062733 | 2.28 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr4_+_135899678 | 2.26 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr14_+_34395845 | 2.25 |
ENSMUST00000048263.14
|
Wapl
|
WAPL cohesin release factor |
| chr1_-_151304191 | 2.24 |
ENSMUST00000064771.12
|
Swt1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr3_-_100396635 | 2.13 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr9_+_74769166 | 2.10 |
ENSMUST00000056006.11
|
Onecut1
|
one cut domain, family member 1 |
| chr5_-_9047324 | 2.09 |
ENSMUST00000003720.5
|
Crot
|
carnitine O-octanoyltransferase |
| chr8_-_58106057 | 2.00 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr18_+_73706115 | 2.00 |
ENSMUST00000091852.5
|
Mex3c
|
mex3 RNA binding family member C |
| chr6_+_91955820 | 2.00 |
ENSMUST00000089334.9
|
Fgd5
|
FYVE, RhoGEF and PH domain containing 5 |
| chr9_+_118435755 | 1.98 |
ENSMUST00000044165.14
|
Itga9
|
integrin alpha 9 |
| chr1_-_171854818 | 1.98 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr9_-_44792575 | 1.96 |
ENSMUST00000114689.8
ENSMUST00000002095.11 ENSMUST00000128768.3 |
Kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr7_+_89779564 | 1.94 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_-_50294652 | 1.93 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr4_-_129121676 | 1.91 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr18_+_89215543 | 1.87 |
ENSMUST00000037142.13
|
Cd226
|
CD226 antigen |
| chr12_-_70033732 | 1.83 |
ENSMUST00000021467.8
|
Sav1
|
salvador family WW domain containing 1 |
| chr4_+_97665843 | 1.82 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chrX_+_13147209 | 1.81 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr9_-_114393406 | 1.81 |
ENSMUST00000111816.3
|
Trim71
|
tripartite motif-containing 71 |
| chr9_-_116004265 | 1.81 |
ENSMUST00000061101.12
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr5_-_143717970 | 1.78 |
ENSMUST00000053287.6
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr15_+_80595486 | 1.74 |
ENSMUST00000067689.9
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr3_-_105708632 | 1.70 |
ENSMUST00000090678.11
|
Rap1a
|
RAS-related protein 1a |
| chr2_-_157046386 | 1.67 |
ENSMUST00000029170.8
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr2_-_62242562 | 1.65 |
ENSMUST00000047812.8
|
Dpp4
|
dipeptidylpeptidase 4 |
| chr4_+_124696336 | 1.63 |
ENSMUST00000138807.8
ENSMUST00000030723.3 |
Mtf1
|
metal response element binding transcription factor 1 |
| chr15_+_6737853 | 1.62 |
ENSMUST00000061656.8
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr4_-_43562397 | 1.60 |
ENSMUST00000030187.14
|
Tln1
|
talin 1 |
| chr10_+_43354807 | 1.59 |
ENSMUST00000167488.9
|
Bend3
|
BEN domain containing 3 |
| chr9_+_51958453 | 1.57 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr2_-_63928339 | 1.56 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr4_+_106173384 | 1.54 |
ENSMUST00000165709.8
ENSMUST00000094933.5 |
Usp24
|
ubiquitin specific peptidase 24 |
| chr1_-_181039509 | 1.53 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr11_+_68322945 | 1.49 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr10_+_128212953 | 1.48 |
ENSMUST00000014642.10
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr11_-_51891259 | 1.43 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr12_-_87346479 | 1.42 |
ENSMUST00000125733.8
|
Ism2
|
isthmin 2 |
| chr5_+_147367237 | 1.37 |
ENSMUST00000176600.8
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr12_-_98703664 | 1.37 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr7_-_144493560 | 1.34 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chrX_-_161671421 | 1.33 |
ENSMUST00000033723.4
|
Syap1
|
synapse associated protein 1 |
| chr5_-_96309849 | 1.29 |
ENSMUST00000155901.8
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr5_+_97145533 | 1.29 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr5_+_128677863 | 1.29 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10 |
| chr5_-_72661776 | 1.28 |
ENSMUST00000175766.8
ENSMUST00000177290.8 ENSMUST00000176974.2 ENSMUST00000167460.9 |
Corin
|
corin, serine peptidase |
| chr11_-_72302520 | 1.26 |
ENSMUST00000108500.8
ENSMUST00000050226.7 |
Smtnl2
|
smoothelin-like 2 |
| chr4_-_53159885 | 1.26 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr14_-_104705420 | 1.25 |
ENSMUST00000053016.10
|
Pou4f1
|
POU domain, class 4, transcription factor 1 |
| chr10_-_116417333 | 1.22 |
ENSMUST00000218744.2
ENSMUST00000105267.8 ENSMUST00000105265.8 ENSMUST00000167706.8 ENSMUST00000168036.8 ENSMUST00000169921.8 ENSMUST00000020374.6 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr10_+_34359395 | 1.22 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr6_-_84570890 | 1.20 |
ENSMUST00000168003.9
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr11_-_49603501 | 1.19 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr16_-_16377982 | 1.19 |
ENSMUST00000161861.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_+_79883885 | 1.16 |
ENSMUST00000163272.2
ENSMUST00000017692.15 |
Suz12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr3_+_65435825 | 1.15 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr4_+_141028539 | 1.14 |
ENSMUST00000006614.3
|
Epha2
|
Eph receptor A2 |
| chr3_+_16237371 | 1.13 |
ENSMUST00000108345.9
ENSMUST00000191774.6 ENSMUST00000108346.5 |
Ythdf3
|
YTH N6-methyladenosine RNA binding protein 3 |
| chr3_-_142587678 | 1.12 |
ENSMUST00000043812.15
|
Pkn2
|
protein kinase N2 |
| chr10_+_69932930 | 1.07 |
ENSMUST00000147545.8
|
Ccdc6
|
coiled-coil domain containing 6 |
| chr19_-_47680528 | 1.06 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr7_+_122723365 | 1.04 |
ENSMUST00000205514.2
ENSMUST00000094053.7 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr18_+_32296126 | 1.04 |
ENSMUST00000096575.5
|
Map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr12_-_69245191 | 1.03 |
ENSMUST00000021356.6
|
Dnaaf2
|
dynein, axonemal assembly factor 2 |
| chr15_+_95688712 | 1.03 |
ENSMUST00000071874.8
|
Ano6
|
anoctamin 6 |
| chr12_+_86407886 | 1.03 |
ENSMUST00000116402.10
|
Esrrb
|
estrogen related receptor, beta |
| chr15_-_57998443 | 1.01 |
ENSMUST00000038194.5
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr1_+_160733942 | 1.01 |
ENSMUST00000161609.8
|
Rc3h1
|
RING CCCH (C3H) domains 1 |
| chr15_+_99476935 | 1.01 |
ENSMUST00000023752.6
|
Aqp2
|
aquaporin 2 |
| chr19_+_18648088 | 1.00 |
ENSMUST00000025632.11
|
Carnmt1
|
carnosine N-methyltransferase 1 |
| chr5_+_88868714 | 1.00 |
ENSMUST00000113229.8
ENSMUST00000006424.8 |
Mob1b
|
MOB kinase activator 1B |
| chr14_+_28226697 | 0.99 |
ENSMUST00000063465.12
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr11_-_86248395 | 0.97 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr6_-_127128007 | 0.96 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chrX_+_104807868 | 0.96 |
ENSMUST00000033581.4
|
Fgf16
|
fibroblast growth factor 16 |
| chr18_-_61533434 | 0.94 |
ENSMUST00000063307.6
ENSMUST00000075299.13 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr16_-_4377640 | 0.94 |
ENSMUST00000005862.9
|
Tfap4
|
transcription factor AP4 |
| chr10_-_7656635 | 0.93 |
ENSMUST00000124838.2
ENSMUST00000039763.14 |
Ginm1
|
glycoprotein integral membrane 1 |
| chr14_+_30201569 | 0.92 |
ENSMUST00000022535.9
ENSMUST00000223658.2 |
Dcp1a
|
decapping mRNA 1A |
| chr2_-_150531280 | 0.92 |
ENSMUST00000046095.10
|
Vsx1
|
visual system homeobox 1 |
| chr14_+_36776775 | 0.91 |
ENSMUST00000120052.2
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr13_-_34529157 | 0.91 |
ENSMUST00000040336.12
|
Slc22a23
|
solute carrier family 22, member 23 |
| chr1_+_133291302 | 0.91 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr13_-_56283331 | 0.91 |
ENSMUST00000045788.9
ENSMUST00000016081.13 |
Macroh2a1
|
macroH2A.1 histone |
| chr13_+_43551851 | 0.88 |
ENSMUST00000071926.5
ENSMUST00000222499.2 |
Nol7
|
nucleolar protein 7 |
| chr18_+_35962832 | 0.88 |
ENSMUST00000060722.8
|
Cxxc5
|
CXXC finger 5 |
| chr15_+_12205095 | 0.87 |
ENSMUST00000038172.16
|
Mtmr12
|
myotubularin related protein 12 |
| chr4_-_133615075 | 0.86 |
ENSMUST00000003741.16
ENSMUST00000105894.11 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr10_-_128727542 | 0.86 |
ENSMUST00000026408.7
|
Gdf11
|
growth differentiation factor 11 |
| chr16_+_35803674 | 0.85 |
ENSMUST00000004054.13
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr13_-_25015392 | 0.84 |
ENSMUST00000006900.7
|
Acot13
|
acyl-CoA thioesterase 13 |
| chr13_-_38178059 | 0.84 |
ENSMUST00000225319.2
ENSMUST00000225246.2 ENSMUST00000021864.8 |
Ssr1
|
signal sequence receptor, alpha |
| chr1_-_152642032 | 0.83 |
ENSMUST00000111859.8
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr14_-_21102487 | 0.83 |
ENSMUST00000154460.8
ENSMUST00000130291.8 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr2_+_131028861 | 0.83 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr3_+_88857929 | 0.82 |
ENSMUST00000186583.7
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr15_+_37233280 | 0.79 |
ENSMUST00000161405.8
ENSMUST00000022895.15 ENSMUST00000161532.2 |
Grhl2
|
grainyhead like transcription factor 2 |
| chr5_-_67585137 | 0.79 |
ENSMUST00000169190.5
|
Bend4
|
BEN domain containing 4 |
| chr1_+_93406686 | 0.78 |
ENSMUST00000027495.15
ENSMUST00000136182.8 ENSMUST00000131175.9 ENSMUST00000153826.8 ENSMUST00000129211.8 ENSMUST00000179353.8 ENSMUST00000172165.8 ENSMUST00000168776.8 |
Septin2
Septin2
|
septin 2 septin 2 |
| chr7_-_127494750 | 0.77 |
ENSMUST00000033074.8
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr11_-_46280336 | 0.75 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr12_+_70499869 | 0.74 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr11_-_95896721 | 0.74 |
ENSMUST00000013559.3
|
Igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr11_-_4544751 | 0.74 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr15_-_50753061 | 0.74 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr11_+_4207557 | 0.73 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr18_+_70701260 | 0.72 |
ENSMUST00000074058.11
ENSMUST00000114946.4 |
Mbd2
|
methyl-CpG binding domain protein 2 |
| chr6_-_37419030 | 0.71 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr8_-_86281946 | 0.69 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr8_+_47192767 | 0.67 |
ENSMUST00000034041.9
ENSMUST00000208507.2 ENSMUST00000207105.2 |
Irf2
|
interferon regulatory factor 2 |
| chr6_+_72391283 | 0.66 |
ENSMUST00000065906.9
|
Ggcx
|
gamma-glutamyl carboxylase |
| chr2_-_33358513 | 0.65 |
ENSMUST00000113156.2
ENSMUST00000028125.12 ENSMUST00000126442.2 |
Zbtb43
|
zinc finger and BTB domain containing 43 |
| chr7_-_74204222 | 0.64 |
ENSMUST00000134539.2
ENSMUST00000026897.14 ENSMUST00000098371.9 |
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr4_-_118148537 | 0.62 |
ENSMUST00000049074.13
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr14_-_8457069 | 0.60 |
ENSMUST00000022257.4
|
Atxn7
|
ataxin 7 |
| chr9_-_101128976 | 0.60 |
ENSMUST00000075941.12
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr3_+_90161470 | 0.59 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr10_+_93476903 | 0.59 |
ENSMUST00000020204.5
|
Ntn4
|
netrin 4 |
| chr12_-_85386120 | 0.59 |
ENSMUST00000040992.8
|
Nek9
|
NIMA (never in mitosis gene a)-related expressed kinase 9 |
| chr1_+_138891447 | 0.59 |
ENSMUST00000168527.8
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr9_-_104140099 | 0.58 |
ENSMUST00000035170.13
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr1_+_6284823 | 0.58 |
ENSMUST00000027040.13
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr2_+_72306503 | 0.58 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr11_+_75422516 | 0.57 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr1_+_139349912 | 0.55 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr8_+_88999031 | 0.55 |
ENSMUST00000169037.9
|
Adcy7
|
adenylate cyclase 7 |
| chr2_+_173501553 | 0.55 |
ENSMUST00000029024.10
ENSMUST00000109110.4 |
Rab22a
|
RAB22A, member RAS oncogene family |
| chr6_+_134897364 | 0.55 |
ENSMUST00000067327.11
ENSMUST00000003115.9 |
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr5_-_140375010 | 0.55 |
ENSMUST00000031539.12
|
Snx8
|
sorting nexin 8 |
| chr7_+_79944198 | 0.54 |
ENSMUST00000163812.9
ENSMUST00000047558.14 ENSMUST00000174199.8 ENSMUST00000173824.8 ENSMUST00000174172.8 |
Prc1
|
protein regulator of cytokinesis 1 |
| chr13_+_55357585 | 0.53 |
ENSMUST00000224973.2
ENSMUST00000099490.3 |
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr19_-_28657477 | 0.53 |
ENSMUST00000162022.8
ENSMUST00000112612.9 |
Glis3
|
GLIS family zinc finger 3 |
| chr3_+_88439616 | 0.52 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr19_-_60569323 | 0.52 |
ENSMUST00000111460.5
ENSMUST00000166712.9 ENSMUST00000081790.15 |
Cacul1
|
CDK2 associated, cullin domain 1 |
| chr15_-_93173032 | 0.52 |
ENSMUST00000057896.5
ENSMUST00000049484.13 ENSMUST00000230063.2 |
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr7_+_132532841 | 0.51 |
ENSMUST00000106157.8
|
Zranb1
|
zinc finger, RAN-binding domain containing 1 |
| chr2_+_68691778 | 0.49 |
ENSMUST00000028426.9
|
Cers6
|
ceramide synthase 6 |
| chr11_-_93859064 | 0.48 |
ENSMUST00000107844.3
ENSMUST00000170303.2 |
Nme1
Gm20390
|
NME/NM23 nucleoside diphosphate kinase 1 predicted gene 20390 |
| chr3_+_32490300 | 0.48 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr1_+_38037086 | 0.48 |
ENSMUST00000027252.8
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr7_-_120673761 | 0.47 |
ENSMUST00000047194.4
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr6_-_28261881 | 0.46 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr7_+_51528788 | 0.46 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr13_-_96678987 | 0.44 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr7_+_96730763 | 0.43 |
ENSMUST00000004622.7
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr13_-_45155616 | 0.43 |
ENSMUST00000072329.15
|
Dtnbp1
|
dystrobrevin binding protein 1 |
| chr10_+_128295159 | 0.42 |
ENSMUST00000026433.9
ENSMUST00000099131.11 ENSMUST00000105235.10 |
Smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr8_+_11808349 | 0.40 |
ENSMUST00000074856.13
ENSMUST00000098938.9 |
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr9_+_58730695 | 0.40 |
ENSMUST00000034889.10
|
Hcn4
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 4 |
| chr16_-_19525122 | 0.40 |
ENSMUST00000081880.7
|
Lamp3
|
lysosomal-associated membrane protein 3 |
| chr12_-_54909568 | 0.40 |
ENSMUST00000078124.8
|
Cfl2
|
cofilin 2, muscle |
| chr10_+_4561974 | 0.39 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr9_-_42383494 | 0.38 |
ENSMUST00000128959.8
ENSMUST00000066148.12 ENSMUST00000138506.8 |
Tbcel
|
tubulin folding cofactor E-like |
| chr6_-_83418656 | 0.38 |
ENSMUST00000089622.11
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr6_+_85400868 | 0.37 |
ENSMUST00000089578.9
|
Noto
|
notochord homeobox |
| chr6_-_83752749 | 0.37 |
ENSMUST00000014892.8
|
Tex261
|
testis expressed gene 261 |
| chr1_-_157240138 | 0.36 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr2_+_75662511 | 0.36 |
ENSMUST00000047232.14
ENSMUST00000111952.9 |
Agps
|
alkylglycerone phosphate synthase |
| chr13_+_14238361 | 0.36 |
ENSMUST00000129488.8
ENSMUST00000110536.8 ENSMUST00000110534.8 ENSMUST00000039538.15 ENSMUST00000110533.2 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr7_+_116980173 | 0.35 |
ENSMUST00000032892.7
|
Xylt1
|
xylosyltransferase 1 |
| chr11_+_70453806 | 0.35 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr2_-_34262012 | 0.34 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr2_-_19002932 | 0.34 |
ENSMUST00000006912.12
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr19_+_43678109 | 0.34 |
ENSMUST00000081079.6
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr11_-_97078698 | 0.33 |
ENSMUST00000001479.5
|
Kpnb1
|
karyopherin (importin) beta 1 |
| chr17_+_25407352 | 0.33 |
ENSMUST00000039734.12
|
Unkl
|
unkempt family like zinc finger |
| chr4_-_151142351 | 0.32 |
ENSMUST00000030797.4
|
Vamp3
|
vesicle-associated membrane protein 3 |
| chr15_+_40518414 | 0.32 |
ENSMUST00000053467.6
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr2_-_131871725 | 0.31 |
ENSMUST00000028814.15
|
Rassf2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr3_-_87081939 | 0.30 |
ENSMUST00000159976.8
ENSMUST00000107618.9 |
Kirrel
|
kirre like nephrin family adhesion molecule 1 |
| chr10_+_93983844 | 0.29 |
ENSMUST00000105290.9
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.0 | 2.9 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.9 | 5.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.7 | 3.0 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.6 | 1.8 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.6 | 0.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.5 | 4.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.5 | 2.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.5 | 1.4 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.5 | 2.3 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 | 2.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.4 | 5.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.4 | 2.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.4 | 2.4 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.4 | 1.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.4 | 2.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.4 | 4.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 1.4 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.3 | 1.0 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.3 | 1.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 3.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.6 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.3 | 1.9 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 1.3 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.3 | 1.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) sensory system development(GO:0048880) |
| 0.3 | 1.9 | GO:0060369 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.3 | 1.8 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.3 | 4.6 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.3 | 2.6 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 0.8 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.3 | 1.6 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 0.8 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 1.0 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.2 | 0.7 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.7 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 1.6 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 1.2 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.2 | 0.6 | GO:0061723 | glycophagy(GO:0061723) |
| 0.2 | 1.3 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 2.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.2 | 1.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.6 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.2 | 1.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 0.5 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 0.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 2.5 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.9 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 2.4 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 1.0 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 1.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.9 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 2.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.6 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 1.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 4.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.9 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.4 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.1 | GO:0072179 | nephric duct formation(GO:0072179) mesonephric duct formation(GO:0072181) |
| 0.1 | 0.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.4 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.9 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.4 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 2.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 3.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.2 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 1.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.3 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.5 | GO:1904706 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 2.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.8 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.0 | 1.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 2.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 2.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 1.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 2.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.8 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.6 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.6 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 4.7 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.9 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 3.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.7 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.3 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 1.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.7 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 | 0.4 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 2.0 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 1.0 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 3.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 1.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.7 | 2.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.5 | 2.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.4 | 2.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 1.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 1.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 2.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 1.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.0 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 0.5 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.4 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 3.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.0 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 1.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 4.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.6 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 7.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 2.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 3.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 4.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 4.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 6.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 3.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.6 | 1.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.4 | 1.3 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 2.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 1.0 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.3 | 1.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 0.8 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 2.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.9 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 1.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.2 | 5.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 2.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.2 | 2.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 0.7 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 1.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 1.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 0.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.0 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 1.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.6 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 1.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 3.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 2.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 6.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 2.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 1.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 5.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 2.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 7.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.9 | GO:0043027 | protein serine/threonine/tyrosine kinase activity(GO:0004712) cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.7 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 6.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 2.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 6.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 1.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 4.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 4.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 5.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 3.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 1.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 4.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 4.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 4.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 2.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 4.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |