Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AGUGGUU
Z-value: 0.72
miRNA associated with seed AGUGGUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-140-5p
|
MIMAT0000151 |
|
mmu-miR-876-3p
|
MIMAT0004855 |
Activity profile of AGUGGUU motif
Sorted Z-values of AGUGGUU motif
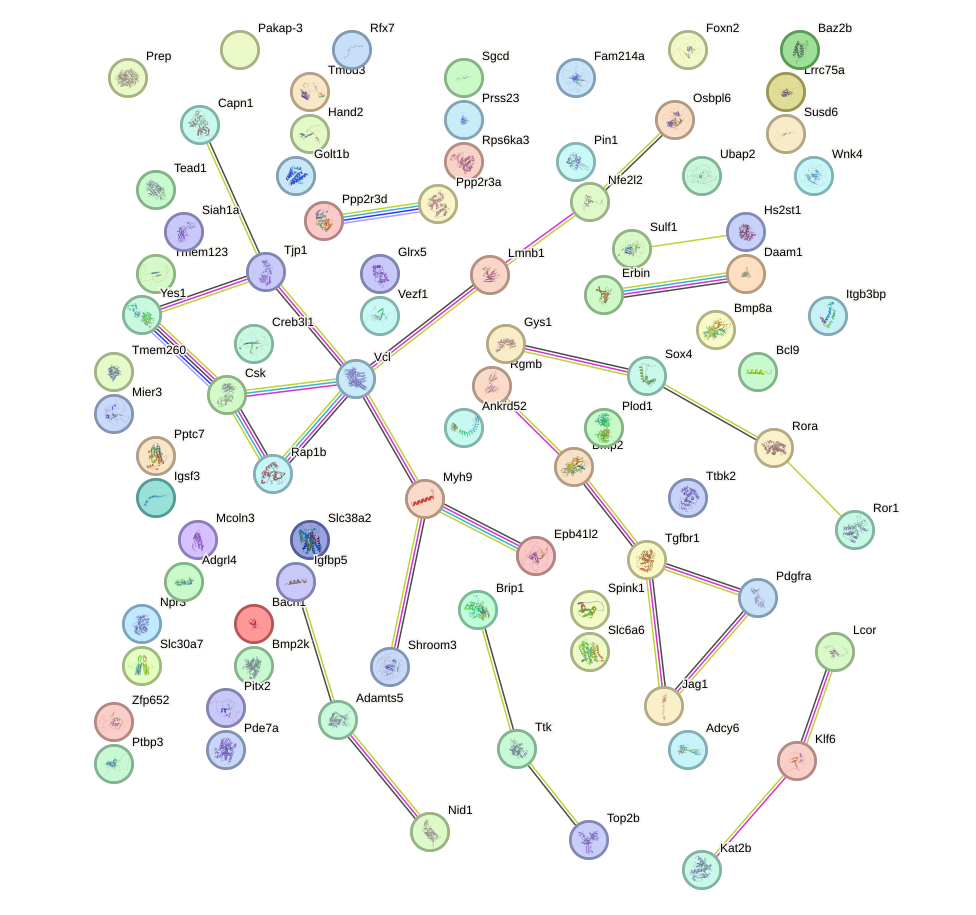
Network of associatons between targets according to the STRING database.
First level regulatory network of AGUGGUU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_57774010 | 4.99 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr18_-_43870622 | 3.97 |
ENSMUST00000025381.4
|
Spink1
|
serine peptidase inhibitor, Kazal type 1 |
| chr2_-_125348305 | 3.94 |
ENSMUST00000028633.13
|
Fbn1
|
fibrillin 1 |
| chr13_-_29137673 | 3.80 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr1_-_72914036 | 3.44 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr3_-_19365431 | 3.33 |
ENSMUST00000099195.10
|
Pde7a
|
phosphodiesterase 7A |
| chr7_+_112278520 | 3.09 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr2_-_136958544 | 2.96 |
ENSMUST00000028735.8
|
Jag1
|
jagged 1 |
| chr4_+_99952988 | 2.90 |
ENSMUST00000039630.6
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr3_+_151143524 | 2.64 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chrX_+_158038778 | 2.52 |
ENSMUST00000126686.8
ENSMUST00000033671.13 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr4_+_57637817 | 2.47 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr15_-_97729341 | 2.43 |
ENSMUST00000079838.14
ENSMUST00000118294.8 |
Hdac7
|
histone deacetylase 7 |
| chr2_-_75534985 | 2.28 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr13_+_5911481 | 2.26 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr9_-_108141105 | 2.26 |
ENSMUST00000166905.8
ENSMUST00000191899.6 |
Dag1
|
dystroglycan 1 |
| chr16_+_87495792 | 2.26 |
ENSMUST00000026703.6
|
Bach1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr10_-_117681864 | 2.25 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr7_+_98484311 | 2.20 |
ENSMUST00000165122.8
ENSMUST00000067495.9 |
Wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr15_-_77726333 | 2.17 |
ENSMUST00000016771.13
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr9_+_7764042 | 2.15 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr11_+_95603494 | 2.13 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr7_-_89166781 | 2.12 |
ENSMUST00000041761.7
|
Prss23
|
protease, serine 23 |
| chr5_+_75312939 | 2.10 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr19_-_6065872 | 2.05 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr7_+_45084257 | 2.00 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr11_+_87959067 | 1.94 |
ENSMUST00000018521.11
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr13_+_13612136 | 1.94 |
ENSMUST00000005532.9
|
Nid1
|
nidogen 1 |
| chr10_+_44943262 | 1.89 |
ENSMUST00000099858.4
|
Prep
|
prolyl endopeptidase |
| chr15_-_98505508 | 1.87 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr11_-_62539284 | 1.77 |
ENSMUST00000057194.9
|
Lrrc75a
|
leucine rich repeat containing 75A |
| chr9_-_85209162 | 1.74 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr6_+_91661034 | 1.74 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr1_+_93406686 | 1.71 |
ENSMUST00000027495.15
ENSMUST00000136182.8 ENSMUST00000131175.9 ENSMUST00000153826.8 ENSMUST00000129211.8 ENSMUST00000179353.8 ENSMUST00000172165.8 ENSMUST00000168776.8 |
Septin2
Septin2
|
septin 2 septin 2 |
| chr4_+_57845240 | 1.65 |
ENSMUST00000102903.8
ENSMUST00000107598.9 |
Pakap
|
paralemmin A kinase anchor protein |
| chr19_+_41471067 | 1.61 |
ENSMUST00000067795.13
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr4_-_59549314 | 1.61 |
ENSMUST00000148331.9
ENSMUST00000030076.12 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr13_+_104424359 | 1.55 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr4_-_148021217 | 1.53 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr3_-_115800989 | 1.51 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr3_-_144275897 | 1.48 |
ENSMUST00000043325.9
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr14_-_6104568 | 1.46 |
ENSMUST00000017629.12
|
Top2b
|
topoisomerase (DNA) II beta |
| chr2_+_133394079 | 1.45 |
ENSMUST00000028836.7
|
Bmp2
|
bone morphogenetic protein 2 |
| chr16_-_85698679 | 1.40 |
ENSMUST00000023611.7
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) |
| chr17_+_53873964 | 1.35 |
ENSMUST00000000724.15
|
Kat2b
|
K(lysine) acetyltransferase 2B |
| chr9_-_101128976 | 1.35 |
ENSMUST00000075941.12
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr18_+_56840813 | 1.33 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr11_+_59197746 | 1.33 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr13_-_104057016 | 1.30 |
ENSMUST00000022222.12
|
Erbin
|
Erbb2 interacting protein |
| chr10_-_13350106 | 1.27 |
ENSMUST00000105545.12
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr12_+_80837284 | 1.25 |
ENSMUST00000220238.2
ENSMUST00000068519.7 |
Susd6
|
sushi domain containing 6 |
| chr3_-_97134680 | 1.21 |
ENSMUST00000046521.14
|
Bcl9
|
B cell CLL/lymphoma 9 |
| chr17_+_74835290 | 1.20 |
ENSMUST00000180037.8
|
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr9_+_72439496 | 1.16 |
ENSMUST00000163401.9
ENSMUST00000093820.10 |
Rfx7
|
regulatory factor X, 7 |
| chr5_+_122422428 | 1.15 |
ENSMUST00000053426.15
|
Pptc7
|
PTC7 protein phosphatase homolog |
| chr2_-_91854844 | 1.15 |
ENSMUST00000028663.5
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr1_+_12762501 | 1.07 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chr9_+_83716736 | 1.05 |
ENSMUST00000185913.7
ENSMUST00000070326.14 ENSMUST00000191484.2 |
Ttk
|
Ttk protein kinase |
| chr13_+_111822712 | 1.05 |
ENSMUST00000109272.9
|
Mier3
|
MIER family member 3 |
| chr5_+_32768515 | 1.04 |
ENSMUST00000202543.4
ENSMUST00000072311.13 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr12_+_104998895 | 0.99 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr9_-_57552844 | 0.98 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr12_-_21467437 | 0.98 |
ENSMUST00000103002.8
ENSMUST00000155480.9 ENSMUST00000135088.9 |
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr4_-_123236596 | 0.98 |
ENSMUST00000102641.10
|
Bmp8a
|
bone morphogenetic protein 8a |
| chr4_+_47353217 | 0.95 |
ENSMUST00000007757.15
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr15_-_11905697 | 0.95 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr2_-_59955995 | 0.90 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr5_+_92957231 | 0.84 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr3_+_146205562 | 0.84 |
ENSMUST00000090031.12
ENSMUST00000118280.2 |
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr14_+_20979466 | 0.81 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr11_-_47270201 | 0.81 |
ENSMUST00000077221.6
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr5_-_143513732 | 0.79 |
ENSMUST00000100489.4
ENSMUST00000080537.14 |
Rac1
|
Rac family small GTPase 1 |
| chr14_+_48683797 | 0.78 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr5_+_97145533 | 0.77 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr11_+_101151394 | 0.74 |
ENSMUST00000103108.8
|
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr17_+_88748139 | 0.74 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr9_+_20563386 | 0.73 |
ENSMUST00000034689.8
|
Pin1
|
protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 |
| chr3_+_101284391 | 0.72 |
ENSMUST00000043983.11
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr15_+_73594965 | 0.72 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr15_-_96597610 | 0.71 |
ENSMUST00000023099.8
|
Slc38a2
|
solute carrier family 38, member 2 |
| chr10_+_25235696 | 0.71 |
ENSMUST00000053748.16
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chrX_+_139565657 | 0.71 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr9_-_75466885 | 0.68 |
ENSMUST00000072232.9
|
Tmod3
|
tropomodulin 3 |
| chr6_-_34854941 | 0.68 |
ENSMUST00000115006.2
ENSMUST00000055097.11 ENSMUST00000115007.9 |
Cyren
|
cell cycle regulator of NHEJ |
| chr9_+_74860335 | 0.67 |
ENSMUST00000170846.8
|
Fam214a
|
family with sequence similarity 214, member A |
| chr4_-_99717359 | 0.67 |
ENSMUST00000146258.2
|
Itgb3bp
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr13_+_81805941 | 0.65 |
ENSMUST00000049055.8
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr16_-_45664664 | 0.65 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr10_+_128212953 | 0.63 |
ENSMUST00000014642.10
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr8_-_87472562 | 0.60 |
ENSMUST00000045296.6
|
Siah1a
|
siah E3 ubiquitin protein ligase 1A |
| chr17_-_16046780 | 0.60 |
ENSMUST00000232638.2
ENSMUST00000170578.3 |
Rgmb
|
repulsive guidance molecule family member B |
| chr4_-_41275091 | 0.59 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr7_-_65020955 | 0.59 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr3_+_128993568 | 0.59 |
ENSMUST00000029657.16
ENSMUST00000106382.11 |
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr12_+_71877838 | 0.59 |
ENSMUST00000223272.2
ENSMUST00000085299.4 |
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr2_+_76236870 | 0.58 |
ENSMUST00000077972.11
ENSMUST00000111929.8 ENSMUST00000111930.9 |
Osbpl6
|
oxysterol binding protein-like 6 |
| chr6_+_142332941 | 0.58 |
ENSMUST00000032372.7
|
Golt1b
|
golgi transport 1B |
| chr3_+_145827410 | 0.55 |
ENSMUST00000039450.5
|
Mcoln3
|
mucolipin 3 |
| chr12_+_56742413 | 0.55 |
ENSMUST00000001538.10
|
Pax9
|
paired box 9 |
| chr9_+_77824646 | 0.54 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr1_+_131838294 | 0.52 |
ENSMUST00000062264.8
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr2_+_31042328 | 0.52 |
ENSMUST00000056433.7
|
Gpr107
|
G protein-coupled receptor 107 |
| chr16_-_76170714 | 0.49 |
ENSMUST00000231585.2
ENSMUST00000121927.8 |
Nrip1
|
nuclear receptor interacting protein 1 |
| chr8_+_36054919 | 0.49 |
ENSMUST00000037666.6
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr7_-_119494918 | 0.49 |
ENSMUST00000059851.14
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr11_-_48717482 | 0.45 |
ENSMUST00000104959.2
|
Gm12184
|
predicted gene 12184 |
| chr18_+_5591864 | 0.45 |
ENSMUST00000025081.13
ENSMUST00000159390.8 |
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr19_+_29499671 | 0.44 |
ENSMUST00000043610.13
|
Ric1
|
RAB6A GEF complex partner 1 |
| chr18_+_53309388 | 0.43 |
ENSMUST00000037850.7
|
Snx2
|
sorting nexin 2 |
| chr8_+_106363141 | 0.43 |
ENSMUST00000005841.16
|
Ctcf
|
CCCTC-binding factor |
| chr11_+_49141339 | 0.42 |
ENSMUST00000101293.11
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr3_+_9315662 | 0.42 |
ENSMUST00000155203.2
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr4_-_53159885 | 0.40 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr9_-_106666329 | 0.40 |
ENSMUST00000046502.7
|
Rad54l2
|
RAD54 like 2 (S. cerevisiae) |
| chr6_+_29348068 | 0.39 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr13_-_18118736 | 0.38 |
ENSMUST00000009003.9
|
Rala
|
v-ral simian leukemia viral oncogene A (ras related) |
| chr6_+_14901343 | 0.37 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr8_-_103512274 | 0.36 |
ENSMUST00000075190.5
|
Cdh11
|
cadherin 11 |
| chr10_-_30476658 | 0.35 |
ENSMUST00000019927.7
|
Trmt11
|
tRNA methyltransferase 11 |
| chr19_+_46587523 | 0.34 |
ENSMUST00000138302.9
ENSMUST00000099376.11 |
Wbp1l
|
WW domain binding protein 1 like |
| chr15_-_51855073 | 0.34 |
ENSMUST00000022927.11
|
Rad21
|
RAD21 cohesin complex component |
| chr11_-_86248395 | 0.33 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr7_-_110682204 | 0.33 |
ENSMUST00000161051.8
ENSMUST00000160132.8 ENSMUST00000162415.9 |
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr1_-_80318197 | 0.32 |
ENSMUST00000163119.8
|
Cul3
|
cullin 3 |
| chr2_+_152873772 | 0.32 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr16_-_46317318 | 0.31 |
ENSMUST00000023335.13
ENSMUST00000023334.15 |
Nectin3
|
nectin cell adhesion molecule 3 |
| chr8_-_25506756 | 0.30 |
ENSMUST00000084032.6
ENSMUST00000207132.2 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr3_+_32583602 | 0.30 |
ENSMUST00000091257.11
|
Mfn1
|
mitofusin 1 |
| chr12_-_69245191 | 0.28 |
ENSMUST00000021356.6
|
Dnaaf2
|
dynein, axonemal assembly factor 2 |
| chr1_+_160733942 | 0.28 |
ENSMUST00000161609.8
|
Rc3h1
|
RING CCCH (C3H) domains 1 |
| chr8_+_108020092 | 0.27 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr18_-_38734389 | 0.27 |
ENSMUST00000025295.8
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr5_+_63969706 | 0.27 |
ENSMUST00000081747.8
ENSMUST00000196575.5 |
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr10_+_62756409 | 0.25 |
ENSMUST00000044977.10
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr3_+_102377234 | 0.24 |
ENSMUST00000035952.5
ENSMUST00000198168.5 ENSMUST00000106925.9 |
Ngf
|
nerve growth factor |
| chr6_-_13677928 | 0.24 |
ENSMUST00000203078.2
ENSMUST00000045235.8 |
Bmt2
|
base methyltransferase of 25S rRNA 2 |
| chr13_+_15638466 | 0.24 |
ENSMUST00000110510.4
|
Gli3
|
GLI-Kruppel family member GLI3 |
| chr7_+_67602565 | 0.24 |
ENSMUST00000005671.10
|
Igf1r
|
insulin-like growth factor I receptor |
| chr14_-_31923803 | 0.24 |
ENSMUST00000226683.2
ENSMUST00000170331.8 ENSMUST00000013845.13 |
Timm23
|
translocase of inner mitochondrial membrane 23 |
| chr13_-_100969878 | 0.23 |
ENSMUST00000067246.6
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr11_-_78642480 | 0.23 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr2_-_119039247 | 0.22 |
ENSMUST00000038439.4
|
Dnajc17
|
DnaJ heat shock protein family (Hsp40) member C17 |
| chr12_-_51738666 | 0.19 |
ENSMUST00000013130.15
ENSMUST00000169503.4 |
Strn3
|
striatin, calmodulin binding protein 3 |
| chr17_-_25493273 | 0.19 |
ENSMUST00000172587.8
ENSMUST00000049911.16 ENSMUST00000173713.8 |
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr1_+_64729603 | 0.18 |
ENSMUST00000114077.8
|
Ccnyl1
|
cyclin Y-like 1 |
| chr14_+_30741082 | 0.18 |
ENSMUST00000112098.11
ENSMUST00000112095.8 ENSMUST00000112106.8 ENSMUST00000146325.8 |
Pbrm1
|
polybromo 1 |
| chr1_+_136552639 | 0.18 |
ENSMUST00000047734.15
ENSMUST00000112046.2 |
Zfp281
|
zinc finger protein 281 |
| chrX_+_52001108 | 0.18 |
ENSMUST00000078944.13
ENSMUST00000101587.10 ENSMUST00000154864.4 |
Phf6
|
PHD finger protein 6 |
| chrX_-_100266032 | 0.17 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr7_-_24919247 | 0.17 |
ENSMUST00000058702.7
|
Dedd2
|
death effector domain-containing DNA binding protein 2 |
| chr8_+_106581719 | 0.17 |
ENSMUST00000040445.9
|
Thap11
|
THAP domain containing 11 |
| chr2_-_156022054 | 0.16 |
ENSMUST00000126992.8
ENSMUST00000146288.8 ENSMUST00000029149.13 ENSMUST00000109587.9 ENSMUST00000109584.8 |
Rbm39
|
RNA binding motif protein 39 |
| chr5_-_51711237 | 0.15 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr3_+_58322119 | 0.14 |
ENSMUST00000099090.7
ENSMUST00000199164.2 |
Tsc22d2
|
TSC22 domain family, member 2 |
| chr19_+_40819682 | 0.13 |
ENSMUST00000025983.13
ENSMUST00000119316.2 |
Ccnj
|
cyclin J |
| chr12_-_108241392 | 0.13 |
ENSMUST00000136175.3
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr1_-_92107971 | 0.13 |
ENSMUST00000186002.3
ENSMUST00000097644.9 |
Hdac4
|
histone deacetylase 4 |
| chr11_-_109364424 | 0.13 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr9_-_59393893 | 0.12 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr5_+_36622342 | 0.12 |
ENSMUST00000031099.4
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr17_+_29768757 | 0.12 |
ENSMUST00000048677.9
ENSMUST00000150388.3 |
Tbc1d22b
Gm28052
|
TBC1 domain family, member 22B predicted gene, 28052 |
| chr10_+_42554888 | 0.11 |
ENSMUST00000040718.6
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr4_-_134014525 | 0.10 |
ENSMUST00000145006.8
ENSMUST00000105877.9 ENSMUST00000127857.2 ENSMUST00000105876.9 |
Pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr15_-_28025920 | 0.10 |
ENSMUST00000090247.7
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr2_-_152673585 | 0.10 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr1_+_40554513 | 0.10 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr4_+_129030710 | 0.09 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr19_-_10181243 | 0.09 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr2_+_145745154 | 0.08 |
ENSMUST00000110000.8
ENSMUST00000002805.14 ENSMUST00000169732.8 ENSMUST00000134759.3 |
Naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr5_+_122296322 | 0.08 |
ENSMUST00000102528.11
ENSMUST00000086294.11 |
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr12_-_91556761 | 0.07 |
ENSMUST00000021345.14
|
Gtf2a1
|
general transcription factor II A, 1 |
| chr9_-_86453862 | 0.05 |
ENSMUST00000070064.11
ENSMUST00000072585.8 |
Pgm3
|
phosphoglucomutase 3 |
| chr2_+_90770742 | 0.05 |
ENSMUST00000005643.14
ENSMUST00000111451.10 ENSMUST00000177642.8 ENSMUST00000068726.13 ENSMUST00000068747.14 |
Celf1
|
CUGBP, Elav-like family member 1 |
| chr6_-_28261881 | 0.05 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr15_-_57939579 | 0.04 |
ENSMUST00000177504.9
ENSMUST00000176076.2 ENSMUST00000177176.8 ENSMUST00000177276.8 ENSMUST00000175805.9 |
Gm29394
Zhx1
|
predicted gene 29394 zinc fingers and homeoboxes 1 |
| chr16_-_75563645 | 0.02 |
ENSMUST00000114244.2
ENSMUST00000046283.16 |
Hspa13
|
heat shock protein 70 family, member 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 1.0 | 3.9 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.9 | 3.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.9 | 5.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.8 | 2.3 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.8 | 2.3 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.7 | 5.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.6 | 1.9 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.6 | 1.7 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.5 | 2.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.5 | 2.2 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.5 | 2.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.5 | 3.0 | GO:0061314 | ciliary body morphogenesis(GO:0061073) Notch signaling involved in heart development(GO:0061314) |
| 0.5 | 2.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.4 | 1.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.4 | 1.4 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.4 | 1.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 1.3 | GO:1904008 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.3 | 1.0 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.3 | 1.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.5 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.3 | 1.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 2.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 1.3 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.2 | 3.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 1.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.5 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 1.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 1.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 2.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.0 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.5 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.6 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 1.7 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 2.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 1.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.3 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.3 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.9 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.8 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 2.9 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 1.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.2 | GO:0022012 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 2.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 0.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.7 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.0 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:1904635 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.0 | 0.7 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 1.0 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.2 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.4 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.7 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.2 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 2.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.9 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.5 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.6 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 2.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0014854 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.5 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.5 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.7 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.8 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 3.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 2.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 3.1 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 3.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.5 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 1.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.0 | 0.3 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 3.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 2.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 10.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.7 | 2.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.6 | 1.7 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.5 | 4.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 3.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 1.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 2.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 5.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 2.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 3.1 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.4 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 1.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 2.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 3.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.8 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 3.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 3.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 4.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 2.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 2.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 1.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.2 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 4.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 3.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 3.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 3.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 3.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 3.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 4.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 2.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |