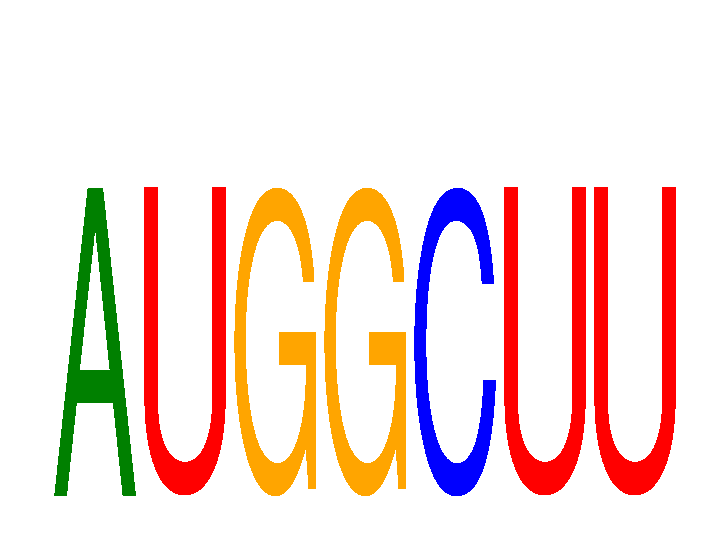Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for AUGGCUU
Z-value: 1.77
miRNA associated with seed AUGGCUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-135a-5p
|
MIMAT0000147 |
|
mmu-miR-135b-5p
|
MIMAT0000612 |
Activity profile of AUGGCUU motif
Sorted Z-values of AUGGCUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AUGGCUU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_108697857 | 34.37 |
ENSMUST00000129040.2
ENSMUST00000046892.10 |
Cplx1
|
complexin 1 |
| chr4_-_87148672 | 24.46 |
ENSMUST00000107157.9
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr7_-_74959010 | 18.39 |
ENSMUST00000165175.8
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr13_+_54519161 | 17.97 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr8_+_96429665 | 16.97 |
ENSMUST00000073139.14
ENSMUST00000080666.8 ENSMUST00000212160.2 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr1_-_173195236 | 16.65 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr2_+_65451100 | 16.34 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr11_+_42310557 | 16.22 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr10_-_108846816 | 15.76 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr1_-_193052568 | 15.54 |
ENSMUST00000016323.11
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr6_-_136150076 | 15.35 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr1_+_34840785 | 15.08 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr5_+_130477642 | 13.99 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr8_+_14145848 | 13.81 |
ENSMUST00000152652.8
ENSMUST00000133298.8 |
Dlgap2
|
DLG associated protein 2 |
| chr8_-_69636825 | 13.64 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr6_-_18514801 | 13.42 |
ENSMUST00000090601.12
|
Cttnbp2
|
cortactin binding protein 2 |
| chr15_-_43034205 | 13.25 |
ENSMUST00000063492.8
ENSMUST00000226810.2 |
Rspo2
|
R-spondin 2 |
| chrX_+_92698469 | 13.05 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr14_-_68362284 | 12.59 |
ENSMUST00000111089.8
ENSMUST00000022638.6 |
Nefm
|
neurofilament, medium polypeptide |
| chr11_+_69217078 | 12.34 |
ENSMUST00000018614.3
|
Kcnab3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr1_-_154602102 | 12.29 |
ENSMUST00000187541.7
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr13_+_117738972 | 12.16 |
ENSMUST00000006991.9
|
Hcn1
|
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
| chr11_+_7013422 | 11.61 |
ENSMUST00000020706.5
|
Adcy1
|
adenylate cyclase 1 |
| chr5_-_103247920 | 11.47 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr5_+_113638313 | 11.30 |
ENSMUST00000094452.4
|
Wscd2
|
WSC domain containing 2 |
| chr9_-_70048766 | 11.09 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr5_+_71857261 | 11.05 |
ENSMUST00000031122.9
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr1_-_22031718 | 11.05 |
ENSMUST00000029667.13
ENSMUST00000173058.8 ENSMUST00000173404.2 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr7_+_43959637 | 10.79 |
ENSMUST00000107938.8
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chrX_+_40490005 | 10.64 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr2_-_136229849 | 10.49 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr6_+_55428963 | 10.43 |
ENSMUST00000070736.12
ENSMUST00000070756.12 ENSMUST00000166962.8 |
Adcyap1r1
|
adenylate cyclase activating polypeptide 1 receptor 1 |
| chr17_+_55752485 | 10.39 |
ENSMUST00000025000.4
|
St6gal2
|
beta galactoside alpha 2,6 sialyltransferase 2 |
| chr1_+_134637031 | 10.36 |
ENSMUST00000121990.2
|
Syt2
|
synaptotagmin II |
| chr13_-_52685305 | 10.31 |
ENSMUST00000057442.8
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr18_-_43820759 | 10.16 |
ENSMUST00000082254.8
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr9_-_108067552 | 9.97 |
ENSMUST00000035208.14
|
Bsn
|
bassoon |
| chr4_+_21931290 | 9.85 |
ENSMUST00000029908.8
|
Faxc
|
failed axon connections homolog |
| chr2_+_83642910 | 9.46 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chrX_-_74621828 | 9.33 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr7_+_29003382 | 9.05 |
ENSMUST00000049977.13
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr10_-_89093441 | 8.68 |
ENSMUST00000182341.8
ENSMUST00000182613.8 |
Ano4
|
anoctamin 4 |
| chr7_+_44033520 | 8.58 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr14_+_27344385 | 8.55 |
ENSMUST00000210135.2
ENSMUST00000090302.6 ENSMUST00000211087.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr7_+_90739904 | 8.24 |
ENSMUST00000107196.10
ENSMUST00000074273.10 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr3_-_110050312 | 8.22 |
ENSMUST00000156177.9
|
Ntng1
|
netrin G1 |
| chr15_+_39061612 | 7.93 |
ENSMUST00000082054.12
ENSMUST00000227243.2 ENSMUST00000042917.10 |
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr17_+_93506590 | 7.91 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr2_-_167032068 | 7.84 |
ENSMUST00000059826.10
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr11_+_67477347 | 7.84 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr6_+_107506678 | 7.81 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr6_+_8948608 | 7.80 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr1_-_22845124 | 7.79 |
ENSMUST00000115273.10
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr1_-_125839897 | 7.69 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr9_-_95632387 | 7.67 |
ENSMUST00000189137.7
ENSMUST00000053785.10 |
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr13_+_83652352 | 7.64 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr8_-_124586159 | 7.63 |
ENSMUST00000034452.12
|
Ccsap
|
centriole, cilia and spindle associated protein |
| chr18_+_23548455 | 7.59 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr11_-_119438569 | 7.40 |
ENSMUST00000026670.5
|
Nptx1
|
neuronal pentraxin 1 |
| chr1_+_166081664 | 7.34 |
ENSMUST00000111416.7
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr9_+_45029080 | 7.34 |
ENSMUST00000170998.9
ENSMUST00000093855.4 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr2_+_55327110 | 7.34 |
ENSMUST00000112633.3
ENSMUST00000112632.2 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr7_+_24181416 | 7.30 |
ENSMUST00000068023.8
|
Cadm4
|
cell adhesion molecule 4 |
| chr2_-_79959178 | 7.26 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr9_+_108685555 | 7.15 |
ENSMUST00000035218.9
ENSMUST00000195323.2 ENSMUST00000194819.2 |
Nckipsd
|
NCK interacting protein with SH3 domain |
| chr14_+_66581818 | 7.09 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chr3_+_153679073 | 6.94 |
ENSMUST00000089948.6
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr3_-_84212069 | 6.93 |
ENSMUST00000107692.8
|
Trim2
|
tripartite motif-containing 2 |
| chr19_-_18978463 | 6.90 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr8_-_17585263 | 6.79 |
ENSMUST00000082104.7
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr12_+_29578354 | 6.77 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr10_+_7465555 | 6.77 |
ENSMUST00000134346.8
ENSMUST00000019931.12 ENSMUST00000130590.8 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr2_+_174485327 | 6.76 |
ENSMUST00000059452.6
|
Zfp831
|
zinc finger protein 831 |
| chr16_-_94798509 | 6.61 |
ENSMUST00000095873.12
ENSMUST00000099508.4 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr13_-_58056089 | 6.54 |
ENSMUST00000185502.7
ENSMUST00000186271.7 ENSMUST00000185905.2 ENSMUST00000187852.7 |
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
| chr10_-_86334700 | 6.48 |
ENSMUST00000120638.8
|
Syn3
|
synapsin III |
| chr13_-_95386776 | 6.47 |
ENSMUST00000162153.9
ENSMUST00000160957.9 ENSMUST00000159598.2 ENSMUST00000162412.8 |
Pde8b
|
phosphodiesterase 8B |
| chr4_+_125384481 | 6.33 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr17_+_26028059 | 6.29 |
ENSMUST00000045692.9
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr7_-_109092834 | 6.23 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr5_+_25964985 | 5.93 |
ENSMUST00000128727.8
ENSMUST00000088244.6 |
Actr3b
|
ARP3 actin-related protein 3B |
| chr3_+_129326004 | 5.83 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr11_+_74510413 | 5.72 |
ENSMUST00000100866.3
|
Ccdc92b
|
coiled-coil domain containing 92B |
| chr10_-_18619439 | 5.71 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr3_-_80820835 | 5.71 |
ENSMUST00000107743.8
ENSMUST00000029654.15 |
Glrb
|
glycine receptor, beta subunit |
| chr10_-_33500583 | 5.66 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr9_+_45749869 | 5.66 |
ENSMUST00000078111.11
ENSMUST00000034591.11 |
Bace1
|
beta-site APP cleaving enzyme 1 |
| chrX_-_166906307 | 5.60 |
ENSMUST00000112149.9
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr3_+_32871669 | 5.59 |
ENSMUST00000072312.12
ENSMUST00000108228.8 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr3_+_115801106 | 5.54 |
ENSMUST00000029575.12
ENSMUST00000106501.8 |
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr5_-_135963408 | 5.47 |
ENSMUST00000198270.2
ENSMUST00000055808.6 |
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
| chr4_-_25800083 | 5.34 |
ENSMUST00000084770.5
|
Fut9
|
fucosyltransferase 9 |
| chr9_+_8544143 | 5.14 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr9_-_42035560 | 5.06 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr7_-_112968533 | 5.04 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chr6_-_35516700 | 4.70 |
ENSMUST00000201026.2
ENSMUST00000031866.9 |
Mtpn
|
myotrophin |
| chr10_+_69369590 | 4.69 |
ENSMUST00000182884.8
|
Ank3
|
ankyrin 3, epithelial |
| chr10_+_82821304 | 4.50 |
ENSMUST00000040110.8
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr13_+_105580147 | 4.49 |
ENSMUST00000022235.6
|
Htr1a
|
5-hydroxytryptamine (serotonin) receptor 1A |
| chr13_+_41403317 | 4.45 |
ENSMUST00000165561.4
|
Smim13
|
small integral membrane protein 13 |
| chr11_+_94881861 | 4.33 |
ENSMUST00000038696.12
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr3_+_157239988 | 4.31 |
ENSMUST00000029831.16
ENSMUST00000106057.8 |
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr8_-_126625029 | 4.28 |
ENSMUST00000047239.13
ENSMUST00000131127.3 |
Pcnx2
|
pecanex homolog 2 |
| chr10_-_121146940 | 4.21 |
ENSMUST00000064107.7
|
Tbc1d30
|
TBC1 domain family, member 30 |
| chr5_-_122187884 | 4.06 |
ENSMUST00000111752.10
|
Cux2
|
cut-like homeobox 2 |
| chr16_+_37909363 | 3.97 |
ENSMUST00000023507.13
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr14_+_120715855 | 3.96 |
ENSMUST00000062117.14
|
Rap2a
|
RAS related protein 2a |
| chr11_+_101066867 | 3.92 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr2_-_37537224 | 3.92 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr8_-_71229293 | 3.91 |
ENSMUST00000034296.15
|
Pik3r2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chrX_+_72546680 | 3.88 |
ENSMUST00000033744.12
ENSMUST00000088429.8 ENSMUST00000114479.2 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr16_-_28748410 | 3.84 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr19_+_16933471 | 3.79 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr12_-_85017586 | 3.77 |
ENSMUST00000165886.2
ENSMUST00000167448.8 ENSMUST00000043169.14 |
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr15_-_74544409 | 3.77 |
ENSMUST00000023268.14
ENSMUST00000110009.4 |
Arc
|
activity regulated cytoskeletal-associated protein |
| chr2_+_163280375 | 3.75 |
ENSMUST00000109420.10
ENSMUST00000109421.10 ENSMUST00000018087.13 ENSMUST00000137070.2 |
Gdap1l1
|
ganglioside-induced differentiation-associated protein 1-like 1 |
| chr10_+_56253418 | 3.70 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr8_-_61436249 | 3.69 |
ENSMUST00000004430.14
ENSMUST00000110301.2 ENSMUST00000093490.9 |
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr17_-_88105422 | 3.66 |
ENSMUST00000055221.9
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr10_-_127047396 | 3.51 |
ENSMUST00000013970.9
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr3_-_138780937 | 3.48 |
ENSMUST00000098574.9
|
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1 |
| chrX_+_9066105 | 3.47 |
ENSMUST00000069763.3
|
Lancl3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chr1_-_133728779 | 3.47 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr7_+_121666388 | 3.44 |
ENSMUST00000033158.6
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr15_+_101071948 | 3.44 |
ENSMUST00000000544.12
|
Acvr1b
|
activin A receptor, type 1B |
| chr7_-_121306476 | 3.33 |
ENSMUST00000046929.7
|
Usp31
|
ubiquitin specific peptidase 31 |
| chr5_+_144037171 | 3.30 |
ENSMUST00000041804.8
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr6_+_54658609 | 3.29 |
ENSMUST00000190641.7
ENSMUST00000187701.2 |
Mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chrX_-_20816841 | 3.28 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr19_-_32717166 | 3.19 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr15_-_78004211 | 3.17 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr11_-_69692542 | 3.17 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chrX_+_165021897 | 3.16 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr18_+_64387428 | 3.15 |
ENSMUST00000025477.15
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chrX_+_7688528 | 3.13 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr4_+_42916666 | 3.09 |
ENSMUST00000132173.8
ENSMUST00000107975.8 |
Phf24
|
PHD finger protein 24 |
| chr6_-_126717590 | 3.08 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr15_+_100659622 | 3.07 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr7_-_141649003 | 3.06 |
ENSMUST00000039926.10
|
Dusp8
|
dual specificity phosphatase 8 |
| chr1_-_14380418 | 3.02 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_-_154249997 | 2.98 |
ENSMUST00000028991.7
ENSMUST00000109728.8 |
Snta1
|
syntrophin, acidic 1 |
| chr8_-_68427217 | 2.97 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_-_77349909 | 2.96 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr3_+_88523440 | 2.93 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr16_-_35589726 | 2.87 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr18_+_23885390 | 2.80 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_+_47167259 | 2.74 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr9_+_21077010 | 2.72 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr3_-_56091096 | 2.71 |
ENSMUST00000029374.8
|
Nbea
|
neurobeachin |
| chr2_+_37406630 | 2.67 |
ENSMUST00000065441.7
|
Gpr21
|
G protein-coupled receptor 21 |
| chr9_+_3532778 | 2.66 |
ENSMUST00000115733.3
|
Gucy1a2
|
guanylate cyclase 1, soluble, alpha 2 |
| chr5_-_44956981 | 2.64 |
ENSMUST00000070748.10
|
Ldb2
|
LIM domain binding 2 |
| chr8_+_85763534 | 2.63 |
ENSMUST00000093360.12
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr4_-_41569500 | 2.60 |
ENSMUST00000108049.9
ENSMUST00000108052.10 ENSMUST00000108050.2 |
Fam219a
|
family with sequence similarity 219, member A |
| chr5_+_13449276 | 2.49 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr15_-_103248512 | 2.37 |
ENSMUST00000168828.3
|
Zfp385a
|
zinc finger protein 385A |
| chr2_+_156038562 | 2.36 |
ENSMUST00000037401.10
|
Phf20
|
PHD finger protein 20 |
| chr8_-_36716445 | 2.35 |
ENSMUST00000239119.2
ENSMUST00000065297.6 |
Lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr14_-_47059694 | 2.33 |
ENSMUST00000111817.8
ENSMUST00000079314.12 |
Gmfb
|
glia maturation factor, beta |
| chr10_+_111000613 | 2.30 |
ENSMUST00000105275.9
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr4_+_129030710 | 2.29 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr10_-_20600797 | 2.26 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr1_-_171023798 | 2.24 |
ENSMUST00000111332.2
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr8_+_110595216 | 2.23 |
ENSMUST00000179721.8
ENSMUST00000034175.5 |
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chrX_-_142716200 | 2.13 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr14_-_60324265 | 2.12 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr5_-_108022900 | 2.10 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr6_+_140568366 | 2.10 |
ENSMUST00000032359.15
|
Aebp2
|
AE binding protein 2 |
| chr17_+_50816296 | 2.09 |
ENSMUST00000043938.8
|
Plcl2
|
phospholipase C-like 2 |
| chr1_-_105284383 | 1.99 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr2_+_151384403 | 1.97 |
ENSMUST00000044011.12
|
Fkbp1a
|
FK506 binding protein 1a |
| chr5_-_143166476 | 1.89 |
ENSMUST00000049861.11
ENSMUST00000165318.4 |
Rbak
|
RB-associated KRAB zinc finger |
| chr19_+_42158995 | 1.86 |
ENSMUST00000169536.8
ENSMUST00000099443.11 |
Zfyve27
|
zinc finger, FYVE domain containing 27 |
| chr11_+_103664976 | 1.83 |
ENSMUST00000000127.6
|
Wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr1_+_166828982 | 1.82 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr6_-_38230890 | 1.80 |
ENSMUST00000117556.8
ENSMUST00000169256.5 |
D630045J12Rik
|
RIKEN cDNA D630045J12 gene |
| chr7_+_24967094 | 1.79 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chr2_+_154498917 | 1.79 |
ENSMUST00000044277.10
|
Chmp4b
|
charged multivesicular body protein 4B |
| chr15_-_77191079 | 1.75 |
ENSMUST00000171751.10
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr8_+_77626400 | 1.72 |
ENSMUST00000109913.9
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr3_-_94794256 | 1.71 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr1_+_75522902 | 1.70 |
ENSMUST00000124341.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr13_+_16189041 | 1.69 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr5_+_138083345 | 1.67 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr18_-_61310066 | 1.57 |
ENSMUST00000235480.2
ENSMUST00000091884.6 |
Hmgxb3
|
HMG box domain containing 3 |
| chrX_-_56438380 | 1.55 |
ENSMUST00000143310.2
ENSMUST00000098470.9 ENSMUST00000114726.8 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr1_+_37338964 | 1.54 |
ENSMUST00000027287.11
ENSMUST00000140264.8 |
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I |
| chr10_+_83379805 | 1.54 |
ENSMUST00000038388.7
|
Washc4
|
WASH complex subunit 4 |
| chr8_-_110335255 | 1.52 |
ENSMUST00000123605.9
ENSMUST00000143900.8 ENSMUST00000128350.3 |
Dhodh
|
dihydroorotate dehydrogenase |
| chr6_+_63232955 | 1.52 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr13_+_104315301 | 1.52 |
ENSMUST00000022225.12
ENSMUST00000069187.12 |
Trim23
|
tripartite motif-containing 23 |
| chr1_+_92759324 | 1.49 |
ENSMUST00000045970.8
|
Gpc1
|
glypican 1 |
| chr3_-_50398027 | 1.49 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr16_+_32882882 | 1.48 |
ENSMUST00000023497.3
|
Lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr4_+_100926170 | 1.47 |
ENSMUST00000106955.2
ENSMUST00000038463.15 |
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr5_-_124465946 | 1.47 |
ENSMUST00000031344.13
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr7_+_18725170 | 1.42 |
ENSMUST00000059331.9
ENSMUST00000131087.2 |
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr7_+_28092370 | 1.38 |
ENSMUST00000003529.9
|
Paf1
|
Paf1, RNA polymerase II complex component |
| chr9_+_109760528 | 1.38 |
ENSMUST00000035055.15
|
Map4
|
microtubule-associated protein 4 |
| chr9_-_103940247 | 1.37 |
ENSMUST00000035166.12
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 4.1 | 12.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 3.3 | 10.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 3.1 | 15.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 2.7 | 13.3 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 2.2 | 20.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.8 | 15.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 1.6 | 11.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.6 | 22.0 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.6 | 7.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.5 | 16.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 1.5 | 7.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.4 | 8.7 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 1.4 | 4.3 | GO:2000474 | cellular response to morphine(GO:0071315) regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.4 | 5.6 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 1.3 | 4.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 1.2 | 3.7 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 1.2 | 7.4 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 1.2 | 3.7 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.2 | 18.0 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 1.2 | 27.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.2 | 3.5 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.1 | 4.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.1 | 10.8 | GO:0046959 | habituation(GO:0046959) |
| 1.0 | 7.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.0 | 7.8 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.9 | 6.5 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.9 | 5.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.8 | 6.8 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.8 | 2.5 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.8 | 3.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.8 | 4.7 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.8 | 3.8 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.7 | 7.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.7 | 10.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.7 | 3.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.7 | 1.4 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.7 | 3.9 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.6 | 7.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.6 | 5.7 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.6 | 3.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.6 | 5.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.6 | 1.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.6 | 1.8 | GO:0060061 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.6 | 2.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.6 | 53.2 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.6 | 3.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 11.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.5 | 10.4 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.5 | 12.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.5 | 26.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.5 | 9.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.5 | 3.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.5 | 2.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.5 | 1.5 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.5 | 9.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.5 | 6.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.5 | 5.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 1.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.4 | 1.7 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.4 | 2.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.4 | 8.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 5.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.4 | 2.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 4.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 5.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.4 | 3.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 13.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.3 | 4.5 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.3 | 1.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 3.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 2.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.3 | 3.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 7.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 3.0 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 1.0 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 1.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.3 | 2.7 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 4.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 0.9 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 23.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 1.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 9.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.5 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 10.6 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 7.7 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.2 | 3.5 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.2 | 0.9 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 1.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.8 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 1.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.8 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.6 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 | 1.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 3.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 16.9 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.2 | 3.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 6.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 5.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 14.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.2 | 1.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 6.1 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 6.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 5.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 0.8 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 2.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 4.1 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.8 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 3.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 7.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 7.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 2.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.0 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.1 | 1.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 3.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 15.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 1.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 8.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 24.4 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.1 | 5.7 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 1.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 1.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.6 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 2.6 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 5.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 3.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 3.9 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.5 | GO:1990166 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.6 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 2.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.2 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.1 | 1.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 4.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.1 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.5 | GO:2000672 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.3 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 6.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.5 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 1.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 8.5 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 3.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 1.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 2.0 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 4.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.9 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 2.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 3.1 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.7 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.0 | 18.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 6.9 | 34.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 4.1 | 12.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 3.3 | 10.0 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 2.5 | 7.6 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 1.6 | 7.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.5 | 7.6 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 1.4 | 26.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.1 | 3.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 1.0 | 27.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.9 | 23.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 11.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 15.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 1.7 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.5 | 14.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.5 | 10.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 1.4 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.4 | 1.7 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.4 | 1.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 5.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 4.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.4 | 4.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.4 | 14.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 11.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 17.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 5.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 51.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 4.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.3 | 3.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 36.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.3 | 5.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 4.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 3.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 1.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 7.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 3.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 5.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 7.7 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 12.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 16.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 4.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 5.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 40.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 22.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 3.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 5.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 4.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 8.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 5.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 10.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 16.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 12.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 3.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 4.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 5.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 8.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 4.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 4.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 7.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 9.8 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 5.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 4.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 5.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 5.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.4 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 9.6 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.1 | 24.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 3.0 | 12.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 2.6 | 10.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 2.4 | 21.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 2.3 | 11.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.0 | 12.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.9 | 15.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.8 | 5.5 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 1.8 | 10.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.7 | 52.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.7 | 10.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.7 | 10.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 1.7 | 11.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.6 | 14.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.5 | 10.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.5 | 4.5 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 1.5 | 13.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.4 | 6.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.3 | 6.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.2 | 3.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.1 | 5.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.1 | 11.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 1.0 | 16.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.9 | 3.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.9 | 7.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.9 | 5.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.9 | 12.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.8 | 7.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.7 | 5.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.7 | 2.7 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.7 | 2.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.7 | 7.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 12.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.6 | 3.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.6 | 1.7 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.6 | 5.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.5 | 4.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.5 | 3.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.5 | 3.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 7.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.5 | 18.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.5 | 12.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.5 | 5.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 6.8 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 3.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 1.5 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.5 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.4 | 1.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 9.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 8.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 8.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 4.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 1.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 15.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 6.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 8.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 7.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 3.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 7.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 26.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 7.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 4.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 9.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.7 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 2.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 6.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 4.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 2.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 10.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 2.4 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 6.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 5.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 2.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 2.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 2.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 5.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 8.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 18.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 21.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.0 | GO:0016840 | arylesterase activity(GO:0004064) carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.6 | GO:0089720 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 5.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 3.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 27.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.0 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 3.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 6.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 11.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 6.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 9.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 13.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.1 | GO:0043621 | protein self-association(GO:0043621) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 24.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.5 | 11.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 28.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 21.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 3.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 10.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 3.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 7.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 6.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 14.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 7.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 29.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.1 | 27.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.8 | 26.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.7 | 11.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.6 | 10.4 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.6 | 13.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.6 | 28.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 37.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.6 | 11.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 19.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.5 | 14.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 5.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.5 | 5.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.4 | 4.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 6.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 3.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 18.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.3 | 9.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 31.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 3.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 7.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 5.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 6.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 1.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 2.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 8.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 6.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 4.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.4 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |