Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Aire
Z-value: 0.91
Transcription factors associated with Aire
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Aire
|
ENSMUSG00000000731.16 | Aire |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Aire | mm39_v1_chr10_-_77879414_77879445 | 0.01 | 9.1e-01 | Click! |
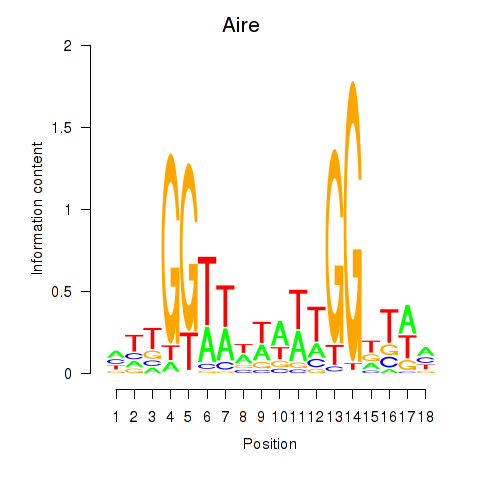
Activity profile of Aire motif
Sorted Z-values of Aire motif
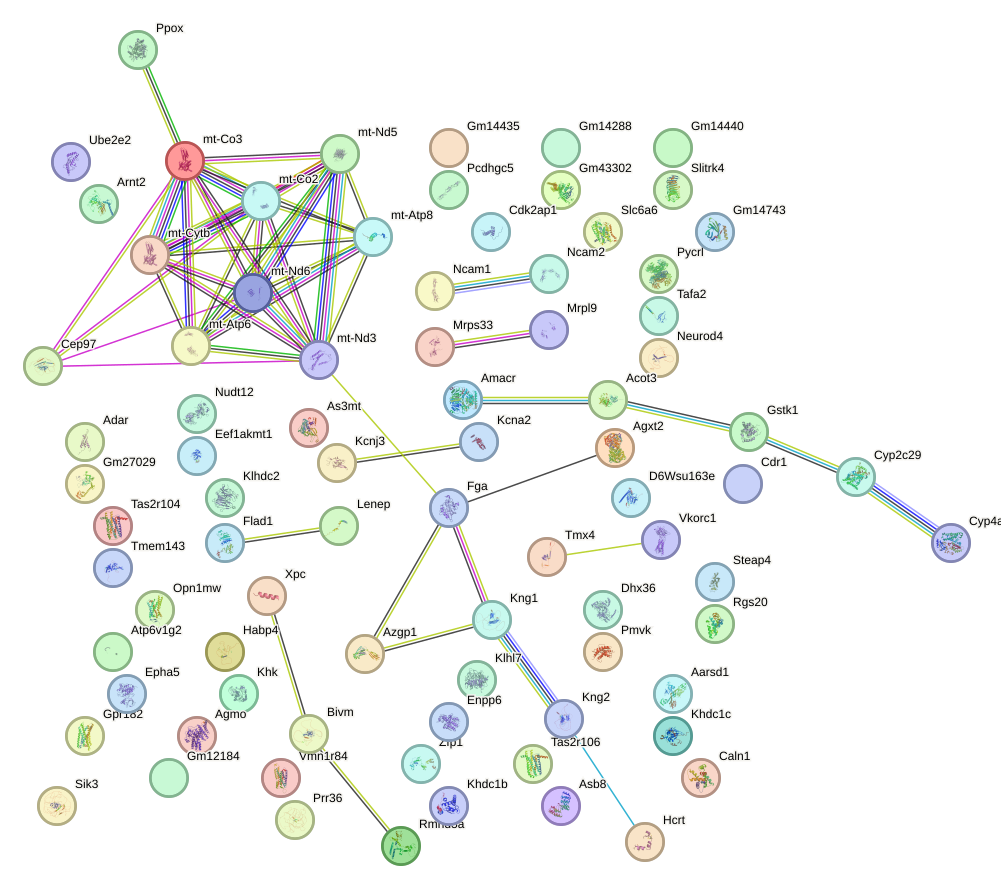
Network of associatons between targets according to the STRING database.
First level regulatory network of Aire
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_60229164 | 25.92 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr16_+_22877000 | 8.01 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr3_+_82933383 | 7.20 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr15_+_10358611 | 5.85 |
ENSMUST00000110541.8
ENSMUST00000110542.8 |
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr19_+_39275518 | 5.83 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr12_+_84098888 | 5.49 |
ENSMUST00000120927.8
ENSMUST00000021653.8 |
Acot3
|
acyl-CoA thioesterase 3 |
| chr15_+_10358643 | 5.00 |
ENSMUST00000022858.8
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr16_-_22847808 | 4.83 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr16_-_22847829 | 4.54 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr5_+_130398261 | 4.06 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chrM_+_7758 | 3.89 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr5_+_137979763 | 3.80 |
ENSMUST00000035390.7
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr10_+_123099945 | 3.73 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chrM_+_7779 | 3.56 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chrM_+_14138 | 3.53 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_+_11735 | 3.49 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_+_8010445 | 3.47 |
ENSMUST00000115421.3
|
Steap4
|
STEAP family member 4 |
| chrM_+_9459 | 3.47 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr2_+_55325931 | 3.27 |
ENSMUST00000067101.10
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr17_+_35455532 | 3.17 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr8_+_47439916 | 3.13 |
ENSMUST00000039840.15
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr10_-_95678748 | 3.03 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr15_+_10981833 | 2.98 |
ENSMUST00000070877.7
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr10_-_95678786 | 2.98 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr9_-_110818679 | 2.94 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chrM_-_14061 | 2.74 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr9_+_109645686 | 2.65 |
ENSMUST00000118732.4
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr10_+_33733706 | 2.61 |
ENSMUST00000218204.2
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr5_+_31079177 | 2.58 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr9_+_109645606 | 2.49 |
ENSMUST00000198988.5
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr18_+_37858753 | 2.45 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr7_-_84059170 | 2.44 |
ENSMUST00000208995.2
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr4_+_115268821 | 2.38 |
ENSMUST00000094887.4
|
Cyp4a12b
|
cytochrome P450, family 4, subfamily a, polypeptide 12B |
| chr7_-_45136235 | 2.27 |
ENSMUST00000210701.2
|
Gm45808
|
predicted gene 45808 |
| chr17_-_59320257 | 2.27 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr10_-_127587576 | 2.25 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr7_+_45546365 | 2.24 |
ENSMUST00000069772.16
ENSMUST00000210503.2 ENSMUST00000209350.2 |
Tmem143
|
transmembrane protein 143 |
| chr7_-_126275529 | 2.21 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr1_-_5140504 | 2.16 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr10_-_106959444 | 2.04 |
ENSMUST00000165067.9
|
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr3_+_107008343 | 1.98 |
ENSMUST00000197470.5
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chrM_+_8603 | 1.93 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr19_+_46695889 | 1.92 |
ENSMUST00000003655.9
|
As3mt
|
arsenite methyltransferase |
| chr7_-_127494750 | 1.92 |
ENSMUST00000033074.8
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr5_-_84565218 | 1.90 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr2_-_134486039 | 1.87 |
ENSMUST00000038228.11
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr11_-_101308441 | 1.81 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr6_+_126916956 | 1.79 |
ENSMUST00000201617.2
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr11_-_49077864 | 1.74 |
ENSMUST00000153999.3
ENSMUST00000066531.13 |
Btnl9
|
butyrophilin-like 9 |
| chr9_-_19163273 | 1.73 |
ENSMUST00000214019.2
ENSMUST00000214267.2 |
Olfr843
|
olfactory receptor 843 |
| chr15_-_76809607 | 1.72 |
ENSMUST00000229865.2
ENSMUST00000229055.2 |
Zfp647
|
zinc finger protein 647 |
| chr5_-_105491795 | 1.72 |
ENSMUST00000171587.2
|
Gbp11
|
guanylate binding protein 11 |
| chr8_-_4267260 | 1.70 |
ENSMUST00000168386.9
|
Prr36
|
proline rich 36 |
| chr1_+_5658716 | 1.69 |
ENSMUST00000160777.8
ENSMUST00000239100.2 ENSMUST00000027038.11 |
Oprk1
|
opioid receptor, kappa 1 |
| chr14_+_3575406 | 1.67 |
ENSMUST00000124353.2
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr16_-_30152708 | 1.59 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chrM_+_7006 | 1.56 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr11_-_115518774 | 1.54 |
ENSMUST00000154623.2
ENSMUST00000106503.10 ENSMUST00000141614.3 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr2_-_134485936 | 1.51 |
ENSMUST00000110120.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr14_+_52964069 | 1.42 |
ENSMUST00000184883.2
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr6_+_42222841 | 1.41 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr13_+_64309675 | 1.38 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chrX_+_73171069 | 1.37 |
ENSMUST00000033771.11
ENSMUST00000101457.10 |
Opn1mw
|
opsin 1 (cone pigments), medium-wave-sensitive (color blindness, deutan) |
| chr6_-_131655849 | 1.32 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr9_-_49479791 | 1.32 |
ENSMUST00000194252.6
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr10_-_130116088 | 1.31 |
ENSMUST00000061571.5
|
Neurod4
|
neurogenic differentiation 4 |
| chr8_-_4267131 | 1.27 |
ENSMUST00000175906.2
|
Prr36
|
proline rich 36 |
| chr14_+_53531798 | 1.26 |
ENSMUST00000103626.3
|
Trav9n-4
|
T cell receptor alpha variable 9N-4 |
| chr8_-_4266912 | 1.26 |
ENSMUST00000177491.8
|
Prr36
|
proline rich 36 |
| chr15_+_89095724 | 1.25 |
ENSMUST00000227951.2
ENSMUST00000226221.2 ENSMUST00000238818.2 ENSMUST00000228284.2 |
Ppp6r2
|
protein phosphatase 6, regulatory subunit 2 |
| chrX_-_63319428 | 1.21 |
ENSMUST00000156121.2
|
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr3_-_89319088 | 1.21 |
ENSMUST00000107429.10
ENSMUST00000129308.9 ENSMUST00000107426.8 ENSMUST00000050398.11 ENSMUST00000162701.2 |
Flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr7_-_12096691 | 1.19 |
ENSMUST00000086228.3
|
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chr6_+_91678630 | 1.18 |
ENSMUST00000205828.2
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr3_+_107008867 | 1.15 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_134485999 | 1.13 |
ENSMUST00000110119.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr6_-_71376277 | 1.12 |
ENSMUST00000149415.2
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr15_-_75793312 | 1.11 |
ENSMUST00000053918.9
|
Pycrl
|
pyrroline-5-carboxylate reductase-like |
| chr8_+_112370088 | 1.10 |
ENSMUST00000077791.8
ENSMUST00000211926.2 |
Zfp1
|
zinc finger protein 1 |
| chr9_+_45924105 | 1.08 |
ENSMUST00000126865.8
|
Sik3
|
SIK family kinase 3 |
| chr17_+_21869545 | 1.02 |
ENSMUST00000232827.2
|
Zfp983
|
zinc finger protein 983 |
| chr16_-_91415873 | 1.00 |
ENSMUST00000143058.2
ENSMUST00000049244.10 ENSMUST00000169982.2 ENSMUST00000133731.2 |
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr15_-_98063493 | 0.99 |
ENSMUST00000143400.8
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr2_+_175125091 | 0.98 |
ENSMUST00000098998.4
|
Gm14440
|
predicted gene 14440 |
| chr2_+_176123622 | 0.98 |
ENSMUST00000172025.2
|
Zfp968-ps
|
zinc finger protein 968, pseudogene |
| chr6_-_131662707 | 0.97 |
ENSMUST00000072404.3
|
Tas2r104
|
taste receptor, type 2, member 104 |
| chr2_-_36975758 | 0.96 |
ENSMUST00000100145.2
|
Olfr361
|
olfactory receptor 361 |
| chr1_+_44159106 | 0.96 |
ENSMUST00000114709.3
ENSMUST00000129068.2 |
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr3_-_62414391 | 0.96 |
ENSMUST00000029336.6
|
Dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr3_+_89366425 | 0.96 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr8_+_112370033 | 0.95 |
ENSMUST00000212072.2
|
Zfp1
|
zinc finger protein 1 |
| chr5_-_124492734 | 0.95 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr6_-_39787813 | 0.95 |
ENSMUST00000114797.2
ENSMUST00000031978.9 |
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr12_+_37292029 | 0.94 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr11_-_58461379 | 0.93 |
ENSMUST00000216758.2
|
Olfr224
|
olfactory receptor 224 |
| chr14_-_57809008 | 0.92 |
ENSMUST00000022518.8
ENSMUST00000239099.2 |
Eef1akmt1
|
EEF1A alpha lysine methyltransferase 1 |
| chr3_+_94350622 | 0.90 |
ENSMUST00000029786.14
ENSMUST00000196143.2 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr6_+_126916919 | 0.89 |
ENSMUST00000032497.7
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr9_+_45924120 | 0.88 |
ENSMUST00000120463.9
ENSMUST00000120247.8 |
Sik3
|
SIK family kinase 3 |
| chr16_-_55755208 | 0.88 |
ENSMUST00000121129.8
ENSMUST00000023270.14 |
Cep97
|
centrosomal protein 97 |
| chr2_+_87854404 | 0.86 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr1_-_171108754 | 0.84 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr3_+_89622323 | 0.83 |
ENSMUST00000098924.9
|
Adar
|
adenosine deaminase, RNA-specific |
| chr5_+_24305577 | 0.83 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr12_+_69343450 | 0.83 |
ENSMUST00000021362.5
|
Klhdc2
|
kelch domain containing 2 |
| chr6_-_91492860 | 0.81 |
ENSMUST00000206476.2
ENSMUST00000032182.5 |
Xpc
|
xeroderma pigmentosum, complementation group C |
| chr6_-_66656668 | 0.80 |
ENSMUST00000071414.2
|
Vmn1r35
|
vomeronasal 1 receptor 35 |
| chr1_+_21438462 | 0.79 |
ENSMUST00000070223.6
|
Khdc1c
|
KH domain containing 1C |
| chrX_-_77289527 | 0.78 |
ENSMUST00000114026.3
|
Gm14743
|
predicted gene 14743 |
| chr18_+_36926929 | 0.74 |
ENSMUST00000001419.10
|
Zmat2
|
zinc finger, matrin type 2 |
| chr11_+_97206542 | 0.71 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr17_+_18801989 | 0.70 |
ENSMUST00000165692.8
|
Vmn2r96
|
vomeronasal 2, receptor 96 |
| chrX_+_77230380 | 0.70 |
ENSMUST00000088198.4
|
Obp1b
|
odorant binding protein IB |
| chr12_-_79030250 | 0.70 |
ENSMUST00000070174.14
|
Tmem229b
|
transmembrane protein 229B |
| chr7_+_20505110 | 0.69 |
ENSMUST00000174538.2
|
Vmn1r112
|
vomeronasal 1 receptor 112 |
| chr6_-_68968278 | 0.69 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr13_+_55593116 | 0.67 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr10_+_52234863 | 0.67 |
ENSMUST00000180473.2
|
Gm26741
|
predicted gene, 26741 |
| chr8_-_85663976 | 0.66 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr14_+_55170152 | 0.65 |
ENSMUST00000037863.6
|
Il25
|
interleukin 25 |
| chrX_+_135418748 | 0.64 |
ENSMUST00000113104.8
ENSMUST00000113105.3 |
Kir3dl1
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 |
| chr19_-_5713728 | 0.64 |
ENSMUST00000169854.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr10_+_80662490 | 0.63 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr16_-_36695166 | 0.61 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chr7_-_102507962 | 0.60 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr1_-_65225617 | 0.59 |
ENSMUST00000186222.7
ENSMUST00000169032.8 ENSMUST00000191459.2 ENSMUST00000188876.7 |
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr7_+_6463510 | 0.57 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chr7_-_47251120 | 0.56 |
ENSMUST00000176369.2
|
Mrgpra3
|
MAS-related GPR, member A3 |
| chr8_+_65399831 | 0.55 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr1_-_65225572 | 0.55 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr6_+_40941688 | 0.54 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr14_-_50521663 | 0.53 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr13_-_105429514 | 0.51 |
ENSMUST00000224011.2
|
Rnf180
|
ring finger protein 180 |
| chr7_-_19398930 | 0.51 |
ENSMUST00000055242.11
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr6_-_58449863 | 0.51 |
ENSMUST00000226390.2
|
Vmn1r31
|
vomeronasal 1 receptor 31 |
| chr7_-_102774821 | 0.50 |
ENSMUST00000211075.3
ENSMUST00000215304.2 ENSMUST00000213281.2 |
Olfr586
|
olfactory receptor 586 |
| chr9_+_104424466 | 0.50 |
ENSMUST00000098443.9
|
Cpne4
|
copine IV |
| chr7_+_19927635 | 0.47 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chr6_-_66614736 | 0.47 |
ENSMUST00000074381.6
|
Vmn1r34
|
vomeronasal 1 receptor 34 |
| chr5_-_121798541 | 0.46 |
ENSMUST00000031412.12
ENSMUST00000111770.2 |
Acad10
|
acyl-Coenzyme A dehydrogenase family, member 10 |
| chr5_-_134589025 | 0.44 |
ENSMUST00000149604.5
|
Syna
|
syncytin a |
| chr7_+_6392535 | 0.43 |
ENSMUST00000207465.2
ENSMUST00000208338.2 |
Zfp28
|
zinc finger protein 28 |
| chr1_+_21419819 | 0.43 |
ENSMUST00000088407.4
|
Khdc1a
|
KH domain containing 1A |
| chr11_+_62349238 | 0.41 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr1_+_173877941 | 0.41 |
ENSMUST00000062665.4
|
Olfr432
|
olfactory receptor 432 |
| chr15_-_76809590 | 0.41 |
ENSMUST00000230677.2
|
Zfp647
|
zinc finger protein 647 |
| chr2_+_85420854 | 0.41 |
ENSMUST00000052307.5
|
Olfr998
|
olfactory receptor 998 |
| chr17_+_18518361 | 0.41 |
ENSMUST00000231938.2
ENSMUST00000079206.8 ENSMUST00000231879.3 |
Vmn2r93
|
vomeronasal 2, receptor 93 |
| chr17_+_20907770 | 0.40 |
ENSMUST00000061756.4
|
Vmn1r226
|
vomeronasal 1 receptor 226 |
| chr2_-_150531280 | 0.40 |
ENSMUST00000046095.10
|
Vsx1
|
visual system homeobox 1 |
| chr1_+_65225886 | 0.39 |
ENSMUST00000097707.5
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr15_-_98063441 | 0.39 |
ENSMUST00000123922.2
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr14_-_56448874 | 0.37 |
ENSMUST00000022757.5
|
Gzmf
|
granzyme F |
| chr14_-_52634836 | 0.34 |
ENSMUST00000214168.2
|
Olfr1511
|
olfactory receptor 1511 |
| chr6_+_56933451 | 0.34 |
ENSMUST00000096612.4
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr4_+_63477018 | 0.34 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chr2_+_83554741 | 0.32 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr17_-_21134625 | 0.30 |
ENSMUST00000077001.3
|
Vmn1r232
|
vomeronasal 1 receptor 232 |
| chr13_-_27574822 | 0.30 |
ENSMUST00000102954.10
ENSMUST00000091678.3 |
Prl2b1
|
prolactin family 2, subfamily b, member 1 |
| chr2_+_172090412 | 0.28 |
ENSMUST00000038532.2
|
Mc3r
|
melanocortin 3 receptor |
| chr2_+_89804937 | 0.26 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr9_+_108883907 | 0.25 |
ENSMUST00000154184.5
|
Shisa5
|
shisa family member 5 |
| chr7_+_25966256 | 0.25 |
ENSMUST00000057123.7
|
Vmn1r184
|
vomeronasal 1 receptor, 184 |
| chr11_+_48977495 | 0.25 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr19_-_5713701 | 0.25 |
ENSMUST00000164304.9
ENSMUST00000237544.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr10_+_129263175 | 0.24 |
ENSMUST00000204529.3
|
Olfr786
|
olfactory receptor 786 |
| chr19_-_5713648 | 0.22 |
ENSMUST00000080824.13
ENSMUST00000237874.2 ENSMUST00000071857.13 ENSMUST00000236464.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr14_+_21531494 | 0.22 |
ENSMUST00000182996.2
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr3_+_92339886 | 0.21 |
ENSMUST00000067102.3
|
Sprr2k
|
small proline-rich protein 2K |
| chrX_-_37665041 | 0.21 |
ENSMUST00000050083.6
ENSMUST00000115118.8 |
Cul4b
|
cullin 4B |
| chr7_+_107679062 | 0.21 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr6_-_58412879 | 0.20 |
ENSMUST00000078890.5
|
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr9_-_39405284 | 0.20 |
ENSMUST00000077757.7
ENSMUST00000215065.3 ENSMUST00000216316.3 |
Olfr44
|
olfactory receptor 44 |
| chr2_-_111320501 | 0.19 |
ENSMUST00000099616.2
|
Olfr1290
|
olfactory receptor 1290 |
| chr9_-_39920878 | 0.19 |
ENSMUST00000216463.3
|
Olfr980
|
olfactory receptor 980 |
| chr2_+_85519775 | 0.19 |
ENSMUST00000054868.2
|
Olfr1008
|
olfactory receptor 1008 |
| chr11_-_66943389 | 0.18 |
ENSMUST00000116363.2
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
| chr7_-_103217838 | 0.17 |
ENSMUST00000214806.2
|
Olfr616
|
olfactory receptor 616 |
| chr7_-_6491875 | 0.17 |
ENSMUST00000036357.7
ENSMUST00000220413.2 |
Olfr1347
|
olfactory receptor 1347 |
| chr10_+_3822667 | 0.16 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr6_-_40976413 | 0.16 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr7_+_6477087 | 0.16 |
ENSMUST00000056144.7
|
Olfr1346
|
olfactory receptor 1346 |
| chr1_+_65225834 | 0.15 |
ENSMUST00000081154.14
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr11_-_83177548 | 0.15 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr1_+_88128323 | 0.14 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr2_-_89159994 | 0.13 |
ENSMUST00000213860.2
ENSMUST00000215679.2 |
Olfr1232
|
olfactory receptor 1232 |
| chr5_-_108943211 | 0.12 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr12_-_36206626 | 0.10 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr7_+_104718676 | 0.09 |
ENSMUST00000098160.2
|
Olfr678
|
olfactory receptor 678 |
| chr9_-_108140925 | 0.05 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr12_+_36207113 | 0.04 |
ENSMUST00000041640.5
|
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr14_+_51351880 | 0.04 |
ENSMUST00000100645.5
|
Eddm3b
|
epididymal protein 3B |
| chr9_+_38164715 | 0.03 |
ENSMUST00000093865.3
|
Olfr143
|
olfactory receptor 143 |
| chr7_+_104963189 | 0.02 |
ENSMUST00000098153.3
|
Olfr689
|
olfactory receptor 689 |
| chr11_-_96002784 | 0.02 |
ENSMUST00000097162.6
ENSMUST00000068686.13 |
Calcoco2
|
calcium binding and coiled-coil domain 2 |
| chrX_-_91059789 | 0.02 |
ENSMUST00000099471.3
ENSMUST00000072269.2 |
Mageb1
|
MAGE family member B1 |
| chr2_-_89969539 | 0.00 |
ENSMUST00000099755.5
|
Olfr32
|
olfactory receptor 32 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 1.8 | 5.5 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 1.8 | 7.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.6 | 2.6 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.6 | 1.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.5 | 3.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.5 | 1.9 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.4 | 3.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.4 | 1.2 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.4 | 1.2 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.4 | 1.5 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.4 | 2.3 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.4 | 1.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 3.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 3.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 1.7 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.3 | 1.3 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.3 | 3.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 5.1 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.3 | 1.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 0.8 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.3 | 0.8 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.3 | 17.4 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.2 | 1.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 3.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.2 | 7.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 9.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 1.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 4.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.0 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.2 | 5.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 0.8 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 0.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.7 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 1.9 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.6 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 0.3 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.1 | 1.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 1.9 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 7.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 3.5 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 0.9 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 2.7 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 4.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.0 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 3.2 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 2.6 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 1.4 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.5 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 3.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 7.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 3.5 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.2 | 1.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 11.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 3.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 4.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 17.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 12.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 5.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.9 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 1.0 | 5.8 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.9 | 2.6 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.6 | 1.9 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.6 | 1.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.4 | 2.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 3.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 3.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 2.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 1.2 | GO:0005369 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.4 | 2.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 3.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 2.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 9.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 0.9 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.3 | 3.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 1.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 1.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 0.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.3 | 5.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 2.3 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.2 | 1.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 1.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.5 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.2 | 4.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 1.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 2.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 3.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 3.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 16.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.7 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 3.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.0 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 2.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 7.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 1.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 1.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 2.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 7.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 2.4 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 1.0 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 2.8 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 7.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 8.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 1.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 3.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |