Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
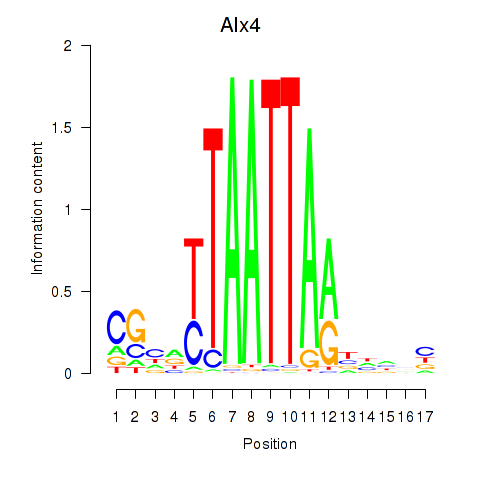
Results for Alx4
Z-value: 0.63
Transcription factors associated with Alx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Alx4
|
ENSMUSG00000040310.13 | Alx4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx4 | mm39_v1_chr2_+_93472657_93472733 | 0.26 | 2.5e-02 | Click! |
Activity profile of Alx4 motif
Sorted Z-values of Alx4 motif
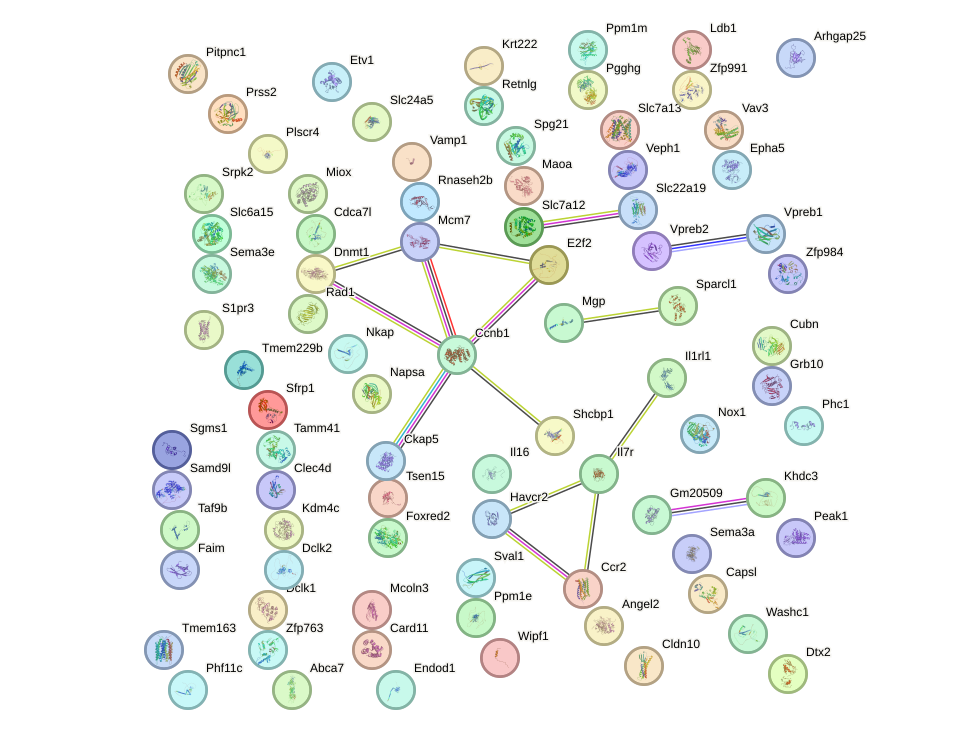
Network of associatons between targets according to the STRING database.
First level regulatory network of Alx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44221791 | 4.84 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr14_+_119025306 | 3.85 |
ENSMUST00000047761.13
ENSMUST00000071546.14 |
Cldn10
|
claudin 10 |
| chr6_-_68609426 | 2.75 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr11_-_106205320 | 2.57 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr2_+_91376650 | 2.54 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr5_+_13448833 | 2.40 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr9_-_20871081 | 2.36 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr19_-_7688628 | 2.27 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr15_+_89218601 | 2.18 |
ENSMUST00000023282.9
|
Miox
|
myo-inositol oxygenase |
| chr10_+_103203552 | 2.16 |
ENSMUST00000179636.3
ENSMUST00000217905.2 ENSMUST00000074204.12 |
Slc6a15
|
solute carrier family 6 (neurotransmitter transporter), member 15 |
| chr4_+_19818718 | 2.03 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr4_+_145311759 | 1.96 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr7_-_24705320 | 1.89 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr7_-_83304698 | 1.88 |
ENSMUST00000145610.8
|
Il16
|
interleukin 16 |
| chr3_-_86827664 | 1.81 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr16_+_45044678 | 1.79 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr19_-_46033353 | 1.79 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr9_-_14292453 | 1.77 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr6_-_87510200 | 1.76 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chrX_-_133012600 | 1.75 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr13_-_100922910 | 1.65 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr3_+_145827410 | 1.63 |
ENSMUST00000039450.5
|
Mcoln3
|
mucolipin 3 |
| chr10_-_62215631 | 1.63 |
ENSMUST00000143236.8
ENSMUST00000133429.8 ENSMUST00000132926.8 ENSMUST00000116238.9 |
Hk1
|
hexokinase 1 |
| chr8_-_4829519 | 1.61 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chrX_-_105264751 | 1.54 |
ENSMUST00000113495.9
|
Taf9b
|
TATA-box binding protein associated factor 9B |
| chr11_-_87249837 | 1.54 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr14_+_51366512 | 1.53 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr11_+_46345784 | 1.53 |
ENSMUST00000109229.2
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr12_-_114012399 | 1.52 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr13_+_51562675 | 1.52 |
ENSMUST00000087978.5
|
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr12_-_114710326 | 1.47 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr4_-_147894245 | 1.46 |
ENSMUST00000105734.10
ENSMUST00000176201.2 |
Zfp984
Gm20707
|
zinc finger protein 984 predicted gene 20707 |
| chr15_-_77840856 | 1.46 |
ENSMUST00000117725.2
ENSMUST00000016696.13 |
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chrX_+_36390430 | 1.44 |
ENSMUST00000016553.5
|
Nkap
|
NFKB activating protein |
| chr3_-_59060907 | 1.43 |
ENSMUST00000196081.5
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr8_+_23901506 | 1.42 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr1_-_171854818 | 1.41 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr9_+_123902143 | 1.41 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr11_+_58839716 | 1.39 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chr3_-_66204228 | 1.38 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr11_+_46345747 | 1.37 |
ENSMUST00000020668.15
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr16_-_16687119 | 1.37 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr6_+_41498716 | 1.37 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr11_-_99134885 | 1.36 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr9_+_92339422 | 1.35 |
ENSMUST00000034941.9
|
Plscr4
|
phospholipid scramblase 4 |
| chr13_+_76727787 | 1.35 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr14_-_104760051 | 1.33 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr6_-_136852792 | 1.32 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr6_+_125192514 | 1.29 |
ENSMUST00000032487.14
ENSMUST00000100942.9 ENSMUST00000063588.11 |
Vamp1
|
vesicle-associated membrane protein 1 |
| chr3_-_86827640 | 1.27 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr16_+_48692976 | 1.27 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr6_+_123239076 | 1.26 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr2_+_124910037 | 1.26 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr6_-_122317484 | 1.23 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr3_+_14545751 | 1.22 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr2_-_13496624 | 1.22 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr6_-_115014777 | 1.21 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr17_-_33252341 | 1.20 |
ENSMUST00000087654.5
|
Zfp763
|
zinc finger protein 763 |
| chr3_+_55369149 | 1.20 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr5_-_104261285 | 1.18 |
ENSMUST00000199947.2
|
Sparcl1
|
SPARC-like 1 |
| chr6_-_3399451 | 1.17 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr12_-_79054050 | 1.17 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr4_+_103000248 | 1.16 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chr4_+_145311722 | 1.16 |
ENSMUST00000105739.8
|
Zfp268
|
zinc finger protein 268 |
| chr1_-_152262425 | 1.16 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chrX_+_16485937 | 1.15 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr6_-_129449739 | 1.14 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr6_-_69282389 | 1.14 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr1_+_40478787 | 1.13 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_-_127605660 | 1.13 |
ENSMUST00000160616.8
|
Tmem163
|
transmembrane protein 163 |
| chr6_+_41928559 | 1.12 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr5_+_136023649 | 1.10 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr12_+_38833454 | 1.10 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr15_-_9529898 | 1.09 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr5_+_13448647 | 1.09 |
ENSMUST00000125629.8
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_-_131001916 | 1.08 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr10_+_79832313 | 1.08 |
ENSMUST00000132517.8
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr17_+_66418525 | 1.08 |
ENSMUST00000072383.14
|
Washc1
|
WASH complex subunit 1 |
| chr5_-_138169253 | 1.06 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr11_-_107228382 | 1.05 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr3_+_109247938 | 1.03 |
ENSMUST00000046864.14
|
Vav3
|
vav 3 oncogene |
| chr1_-_83016152 | 1.02 |
ENSMUST00000164473.2
ENSMUST00000045560.15 |
Slc19a3
|
solute carrier family 19, member 3 |
| chr11_-_11920540 | 1.02 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr5_+_14075281 | 1.02 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr9_+_65368207 | 1.02 |
ENSMUST00000034955.8
ENSMUST00000213957.2 |
Spg21
|
SPG21, maspardin |
| chr7_+_89814713 | 1.02 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr15_+_9436114 | 1.02 |
ENSMUST00000042360.5
ENSMUST00000226688.2 |
Capsl
|
calcyphosine-like |
| chr12_-_75678092 | 1.02 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr4_+_147216495 | 1.01 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991 |
| chr19_-_32173824 | 1.01 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr4_+_74160705 | 0.99 |
ENSMUST00000077851.10
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr6_+_70640233 | 0.99 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr5_-_140986312 | 0.98 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr2_+_87696836 | 0.97 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr2_-_73284262 | 0.97 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_190660689 | 0.96 |
ENSMUST00000066632.14
ENSMUST00000110899.7 |
Angel2
|
angel homolog 2 |
| chr9_+_73009680 | 0.96 |
ENSMUST00000034737.13
ENSMUST00000173734.9 ENSMUST00000167514.2 ENSMUST00000174203.3 |
Khdc3
Gm20509
|
KH domain containing 3, subcortical maternal complex member predicted gene 20509 |
| chr5_-_114911548 | 0.95 |
ENSMUST00000178440.8
ENSMUST00000043283.14 ENSMUST00000112185.9 ENSMUST00000155908.8 |
Git2
|
GIT ArfGAP 2 |
| chr7_+_140521450 | 0.93 |
ENSMUST00000164580.3
ENSMUST00000079403.11 |
Pgghg
|
protein glucosylgalactosylhydroxylysine glucosidase |
| chr5_-_84565218 | 0.92 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr9_-_56151334 | 0.92 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr4_+_135899678 | 0.91 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr6_+_68279392 | 0.91 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr9_+_38738911 | 0.89 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr9_+_98873831 | 0.89 |
ENSMUST00000185472.2
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr14_+_62529924 | 0.89 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr12_+_117807224 | 0.88 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like |
| chr14_-_59602882 | 0.88 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr5_-_23821523 | 0.87 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr4_-_70328659 | 0.87 |
ENSMUST00000144099.8
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr6_+_41511248 | 0.85 |
ENSMUST00000192366.2
ENSMUST00000103286.2 |
Trbj1-3
|
T cell receptor beta joining 1-3 |
| chr11_-_76386190 | 0.85 |
ENSMUST00000108408.9
|
Abr
|
active BCR-related gene |
| chr14_+_73376192 | 0.85 |
ENSMUST00000171070.8
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr5_-_114911432 | 0.85 |
ENSMUST00000112183.8
|
Git2
|
GIT ArfGAP 2 |
| chr6_-_69704122 | 0.85 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr19_+_12647803 | 0.85 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr7_+_19927635 | 0.84 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chr6_+_92793440 | 0.84 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr10_+_29074950 | 0.83 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr15_-_93493758 | 0.82 |
ENSMUST00000048982.11
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr1_-_88205233 | 0.81 |
ENSMUST00000065420.12
ENSMUST00000054674.15 |
Hjurp
|
Holliday junction recognition protein |
| chr5_-_114911509 | 0.81 |
ENSMUST00000086564.11
|
Git2
|
GIT ArfGAP 2 |
| chr5_-_151574620 | 0.81 |
ENSMUST00000038131.10
|
Rfc3
|
replication factor C (activator 1) 3 |
| chr5_+_107645626 | 0.81 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr3_+_90200470 | 0.81 |
ENSMUST00000199754.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr6_+_70648743 | 0.80 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr2_-_144112700 | 0.80 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5 |
| chr9_-_59260713 | 0.79 |
ENSMUST00000026265.8
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr1_+_160898283 | 0.79 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr11_+_35011953 | 0.78 |
ENSMUST00000069837.4
|
Slit3
|
slit guidance ligand 3 |
| chr3_+_87989278 | 0.78 |
ENSMUST00000071812.11
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr4_+_102446883 | 0.78 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr11_-_58059293 | 0.78 |
ENSMUST00000172035.8
ENSMUST00000035604.13 ENSMUST00000102711.9 |
Gemin5
|
gem nuclear organelle associated protein 5 |
| chr17_+_66418598 | 0.78 |
ENSMUST00000116556.4
ENSMUST00000233354.2 |
Washc1
|
WASH complex subunit 1 |
| chr2_-_126342551 | 0.77 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr15_+_80017315 | 0.77 |
ENSMUST00000023050.9
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr1_-_152262339 | 0.77 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr11_+_106107752 | 0.76 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42 |
| chr14_+_15369152 | 0.76 |
ENSMUST00000167923.8
|
Gm3696
|
predicted gene 3696 |
| chr16_-_45544960 | 0.75 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr9_+_118892497 | 0.74 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr3_-_105839980 | 0.74 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr19_-_8796288 | 0.74 |
ENSMUST00000153281.2
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr3_-_113325938 | 0.74 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr5_-_138169509 | 0.74 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr12_-_69939931 | 0.73 |
ENSMUST00000049239.8
ENSMUST00000110570.8 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr5_-_65855511 | 0.73 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr6_-_68887957 | 0.72 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr2_-_111843053 | 0.72 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chrX_-_121307036 | 0.72 |
ENSMUST00000079490.6
|
Nap1l3
|
nucleosome assembly protein 1-like 3 |
| chr6_+_41512010 | 0.72 |
ENSMUST00000103288.2
|
Trbj1-5
|
T cell receptor beta joining 1-5 |
| chr15_-_36794741 | 0.72 |
ENSMUST00000110361.8
ENSMUST00000022894.14 ENSMUST00000110359.2 |
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr10_+_80003612 | 0.72 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr19_+_5524701 | 0.72 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr11_-_29197222 | 0.72 |
ENSMUST00000020754.10
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr6_+_129022843 | 0.71 |
ENSMUST00000032257.10
ENSMUST00000204320.2 |
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F |
| chr14_+_53599724 | 0.71 |
ENSMUST00000196105.2
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr5_+_121601212 | 0.70 |
ENSMUST00000094357.11
ENSMUST00000031405.12 |
Tmem116
|
transmembrane protein 116 |
| chr4_-_136329953 | 0.70 |
ENSMUST00000105847.8
ENSMUST00000116273.9 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr4_+_12906838 | 0.70 |
ENSMUST00000143186.8
ENSMUST00000183345.2 |
Triqk
|
triple QxxK/R motif containing |
| chr3_+_142326363 | 0.69 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr2_-_29677634 | 0.69 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr4_-_106536063 | 0.69 |
ENSMUST00000106772.10
ENSMUST00000135676.2 ENSMUST00000026480.13 |
Ttc4
|
tetratricopeptide repeat domain 4 |
| chr17_-_21110913 | 0.69 |
ENSMUST00000061278.2
|
Vmn1r231
|
vomeronasal 1 receptor 231 |
| chrX_+_56257374 | 0.69 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr2_-_88157559 | 0.68 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr2_-_73490746 | 0.68 |
ENSMUST00000102677.11
|
Chn1
|
chimerin 1 |
| chr18_+_37898633 | 0.67 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr12_-_113790741 | 0.67 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr6_+_57180275 | 0.67 |
ENSMUST00000226892.2
ENSMUST00000227421.2 |
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr5_-_3691453 | 0.65 |
ENSMUST00000140871.2
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr15_-_79658584 | 0.65 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr15_-_66985760 | 0.65 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_+_100900278 | 0.65 |
ENSMUST00000103110.10
ENSMUST00000044721.13 ENSMUST00000168757.9 |
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr9_-_123507937 | 0.65 |
ENSMUST00000040960.13
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr19_-_10807409 | 0.64 |
ENSMUST00000080292.12
|
Cd6
|
CD6 antigen |
| chr7_-_30259025 | 0.64 |
ENSMUST00000043975.11
ENSMUST00000156241.2 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr19_-_10807285 | 0.64 |
ENSMUST00000039043.15
|
Cd6
|
CD6 antigen |
| chr10_+_129493563 | 0.64 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr5_+_3593811 | 0.63 |
ENSMUST00000197082.5
ENSMUST00000115527.8 |
Fam133b
|
family with sequence similarity 133, member B |
| chr9_-_123507847 | 0.63 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr13_+_67961859 | 0.62 |
ENSMUST00000181391.8
ENSMUST00000012725.8 |
Zfp273
|
zinc finger protein 273 |
| chr6_-_66537080 | 0.62 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr12_-_69939762 | 0.62 |
ENSMUST00000110567.8
ENSMUST00000171211.8 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_+_35188888 | 0.62 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr5_+_27022355 | 0.62 |
ENSMUST00000071500.13
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr4_+_146695418 | 0.62 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr9_-_22171180 | 0.61 |
ENSMUST00000086281.5
|
Zfp599
|
zinc finger protein 599 |
| chr2_-_34716199 | 0.61 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr19_-_11582207 | 0.60 |
ENSMUST00000025582.11
|
Ms4a6d
|
membrane-spanning 4-domains, subfamily A, member 6D |
| chr11_+_102080489 | 0.59 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr15_-_79658608 | 0.59 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr2_+_174169539 | 0.59 |
ENSMUST00000133356.8
ENSMUST00000087871.11 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr5_-_108943211 | 0.58 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr4_-_135714465 | 0.58 |
ENSMUST00000105851.9
|
Pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr4_-_43710231 | 0.58 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr8_-_86091946 | 0.57 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr7_-_30523191 | 0.57 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chr7_-_30259253 | 0.57 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr10_-_128462616 | 0.56 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.7 | 2.9 | GO:0002856 | negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.6 | 1.8 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.6 | 2.4 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 1.4 | GO:2000040 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 1.4 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.5 | 1.4 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.4 | 2.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 2.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.4 | 1.6 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.4 | 1.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.3 | 1.4 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.3 | 1.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.3 | 1.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 1.3 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 2.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.3 | 4.8 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.3 | 2.3 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.3 | 0.8 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 1.9 | GO:1904109 | positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.2 | 0.7 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 1.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.2 | 1.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.6 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.8 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 0.8 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 | 1.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 0.7 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.2 | 0.5 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 1.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 2.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 1.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 1.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 1.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 1.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.6 | GO:0002879 | cell surface pattern recognition receptor signaling pathway(GO:0002752) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.4 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 1.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.3 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 1.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.4 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 1.9 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 0.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.9 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 1.2 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 1.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 1.8 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 1.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 1.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.3 | GO:0061738 | mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) |
| 0.1 | 1.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.6 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 0.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.8 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 1.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.7 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.4 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 2.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.3 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 1.0 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 0.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 2.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 1.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.9 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.3 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.7 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.6 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 1.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.8 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 8.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 1.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 2.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 1.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 1.4 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.4 | GO:2001198 | regulation of dendritic cell differentiation(GO:2001198) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.7 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 5.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.8 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.9 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 1.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 1.1 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.3 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:1901750 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 1.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 1.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.4 | 4.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.3 | 2.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 2.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 1.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 0.9 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 0.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 1.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 2.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 3.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 1.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.8 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 3.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 1.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 4.1 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 7.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.4 | 1.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 1.1 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 2.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 3.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 1.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 4.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 1.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 1.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 0.8 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.3 | 1.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 4.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.6 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 2.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 1.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 1.0 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 1.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 2.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 2.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 4.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.5 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.4 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 2.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.1 | 1.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 2.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 5.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 3.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 6.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 5.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 2.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 4.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 1.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 2.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 2.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 3.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |