Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
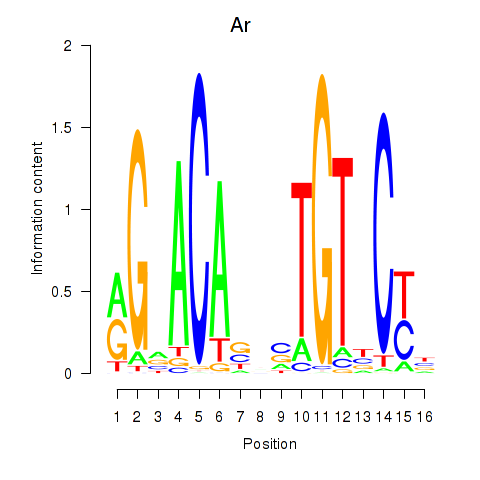
Results for Ar
Z-value: 1.90
Transcription factors associated with Ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ar
|
ENSMUSG00000046532.9 | Ar |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ar | mm39_v1_chrX_+_97192356_97192388 | -0.60 | 3.3e-08 | Click! |
Activity profile of Ar motif
Sorted Z-values of Ar motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ar
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_152626273 | 13.99 |
ENSMUST00000068875.5
|
Apobec4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr2_-_32277773 | 9.99 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr2_+_118998235 | 9.63 |
ENSMUST00000057454.4
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr11_-_69712970 | 8.32 |
ENSMUST00000045771.7
|
Spem1
|
sperm maturation 1 |
| chr6_-_69678271 | 7.95 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr11_+_108286114 | 7.91 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr15_-_85705935 | 7.75 |
ENSMUST00000064370.6
|
Pkdrej
|
polycystin (PKD) family receptor for egg jelly |
| chr9_-_22043083 | 7.71 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr8_+_123912976 | 7.56 |
ENSMUST00000019422.6
|
Dpep1
|
dipeptidase 1 |
| chr17_+_80434874 | 7.46 |
ENSMUST00000039205.11
|
Galm
|
galactose mutarotase |
| chr9_+_7692087 | 7.29 |
ENSMUST00000018767.8
|
Mmp7
|
matrix metallopeptidase 7 |
| chr2_+_24970327 | 7.14 |
ENSMUST00000044078.10
ENSMUST00000114380.9 |
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr15_+_75088445 | 6.93 |
ENSMUST00000055719.8
|
Ly6g2
|
lymphocyte antigen 6 complex, locus G2 |
| chr1_+_13738967 | 6.91 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr10_-_99595498 | 6.75 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr8_+_21624626 | 6.75 |
ENSMUST00000098898.5
|
Defa30
|
defensin, alpha, 30 |
| chr16_-_18880821 | 6.70 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr7_+_30676465 | 6.62 |
ENSMUST00000058093.6
|
Fam187b
|
family with sequence similarity 187, member B |
| chr1_+_172525613 | 6.47 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr4_+_134042423 | 6.42 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr2_-_32278245 | 6.41 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr9_-_22042930 | 6.33 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr9_+_48406706 | 6.32 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr14_-_33996185 | 6.29 |
ENSMUST00000227006.2
|
Shld2
|
shieldin complex subunit 2 |
| chr7_+_79896121 | 6.26 |
ENSMUST00000058266.9
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr6_-_72212547 | 6.25 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr4_-_141345549 | 6.17 |
ENSMUST00000053263.9
|
Tmem82
|
transmembrane protein 82 |
| chr4_-_44710408 | 6.16 |
ENSMUST00000134968.9
ENSMUST00000173821.8 ENSMUST00000174319.8 ENSMUST00000173733.8 ENSMUST00000172866.8 ENSMUST00000165417.9 ENSMUST00000107825.9 ENSMUST00000102932.10 ENSMUST00000107827.9 ENSMUST00000107826.9 ENSMUST00000014174.14 |
Pax5
|
paired box 5 |
| chr7_-_19415301 | 6.12 |
ENSMUST00000150569.9
ENSMUST00000127648.4 ENSMUST00000003071.10 |
Gm44805
Apoc4
|
predicted gene 44805 apolipoprotein C-IV |
| chr1_+_88015524 | 6.12 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr8_-_21946576 | 6.11 |
ENSMUST00000110752.4
|
Defa42
|
defensin, alpha, 42 |
| chr5_-_134944366 | 6.11 |
ENSMUST00000008987.5
|
Cldn13
|
claudin 13 |
| chr2_-_162929732 | 6.04 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr11_-_58343747 | 6.01 |
ENSMUST00000064614.4
|
Lypd9
|
LY6/PLAUR domain containing 9 |
| chr4_-_49408040 | 5.95 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr8_-_22193658 | 5.95 |
ENSMUST00000071886.7
|
Defa39
|
defensin, alpha, 39 |
| chr11_-_75330415 | 5.93 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr11_+_96822213 | 5.91 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr8_+_22224506 | 5.88 |
ENSMUST00000080533.6
|
Defa24
|
defensin, alpha, 24 |
| chr3_+_97536120 | 5.86 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr1_+_88128323 | 5.75 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr17_-_84154173 | 5.70 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr14_+_15137378 | 5.61 |
ENSMUST00000100886.4
|
Gm5797
|
predicted gene 5797 |
| chr7_+_43701714 | 5.51 |
ENSMUST00000079859.7
|
Klk1b27
|
kallikrein 1-related peptidase b27 |
| chr14_+_16343966 | 5.50 |
ENSMUST00000112742.4
ENSMUST00000167913.8 |
Gm3476
|
predicted gene 3476 |
| chr1_-_53226756 | 5.44 |
ENSMUST00000190748.2
ENSMUST00000072235.10 |
1700019A02Rik
|
RIKEN cDNA 1700019A02 gene |
| chr8_-_21586066 | 5.44 |
ENSMUST00000077452.4
|
Defa38
|
defensin, alpha, 38 |
| chr14_-_18152432 | 5.43 |
ENSMUST00000171810.8
ENSMUST00000112779.10 |
Gm3149
|
predicted gene 3149 |
| chr14_+_15695362 | 5.37 |
ENSMUST00000170673.2
ENSMUST00000168790.8 |
Gm8362
|
predicted gene 8362 |
| chr11_-_75330302 | 5.37 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr7_+_118199375 | 5.32 |
ENSMUST00000121744.9
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr8_+_21545063 | 5.28 |
ENSMUST00000098899.4
|
Defa23
|
defensin, alpha, 23 |
| chr7_-_46392403 | 5.28 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr11_-_78313043 | 5.24 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr16_-_97306125 | 5.24 |
ENSMUST00000049721.9
ENSMUST00000231999.2 |
Fam3b
|
family with sequence similarity 3, member B |
| chr1_+_177557380 | 5.23 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr8_+_22145796 | 5.22 |
ENSMUST00000079528.6
|
Defa17
|
defensin, alpha, 17 |
| chr12_-_113790741 | 5.20 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr6_+_124489364 | 5.10 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr6_+_138119851 | 5.07 |
ENSMUST00000125810.2
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr7_+_119125546 | 5.06 |
ENSMUST00000207387.2
ENSMUST00000207813.2 |
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr1_-_167294349 | 5.04 |
ENSMUST00000036643.6
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr12_+_31440842 | 5.01 |
ENSMUST00000167432.8
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr19_+_43770619 | 4.99 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chrX_-_126644166 | 4.97 |
ENSMUST00000052500.6
|
Cldn34c4
|
claudin 34C4 |
| chr10_-_117118226 | 4.95 |
ENSMUST00000092163.9
|
Lyz2
|
lysozyme 2 |
| chr8_+_120301974 | 4.95 |
ENSMUST00000093100.3
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr6_-_68681962 | 4.90 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr5_+_25721059 | 4.89 |
ENSMUST00000045016.9
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr5_-_24760401 | 4.89 |
ENSMUST00000088302.10
|
Iqca1l
|
IQ motif containing with AAA domain 1 like |
| chr1_+_165958036 | 4.88 |
ENSMUST00000166860.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr17_-_84154196 | 4.83 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr12_-_103597663 | 4.77 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr17_-_12894716 | 4.75 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr16_-_18884431 | 4.68 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr7_+_28682253 | 4.67 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr2_+_156681927 | 4.65 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr15_+_7159038 | 4.63 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr2_-_28453374 | 4.58 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr8_+_21777425 | 4.56 |
ENSMUST00000098893.4
|
Defa3
|
defensin, alpha, 3 |
| chr6_+_29279582 | 4.55 |
ENSMUST00000167131.8
|
Fam71f2
|
family with sequence similarity 71, member F2 |
| chr3_+_10431961 | 4.51 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr3_-_98721750 | 4.43 |
ENSMUST00000029463.13
|
Hsd3b6
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 6 |
| chr15_-_82291372 | 4.43 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr5_+_90608751 | 4.41 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr4_+_116565706 | 4.39 |
ENSMUST00000030452.13
ENSMUST00000106462.9 |
Ccdc163
|
coiled-coil domain containing 163 |
| chr11_+_62842019 | 4.37 |
ENSMUST00000035854.4
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr7_+_119125443 | 4.34 |
ENSMUST00000207440.2
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr16_-_97564910 | 4.32 |
ENSMUST00000019386.10
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr3_+_146276147 | 4.30 |
ENSMUST00000199489.5
|
Uox
|
urate oxidase |
| chr8_-_22084524 | 4.29 |
ENSMUST00000110749.4
|
Defa43
|
defensin, alpha, 43 |
| chr1_-_136876902 | 4.27 |
ENSMUST00000195428.2
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chrX_-_101751241 | 4.26 |
ENSMUST00000113610.3
|
Gm9112
|
predicted gene 9112 |
| chr5_-_145521533 | 4.23 |
ENSMUST00000075837.8
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr8_+_84728123 | 4.20 |
ENSMUST00000060357.15
ENSMUST00000239176.2 |
1700067K01Rik
|
RIKEN cDNA 1700067K01 gene |
| chr15_-_102112159 | 4.20 |
ENSMUST00000229252.2
ENSMUST00000229770.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr8_+_118428643 | 4.15 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr7_+_119125426 | 4.14 |
ENSMUST00000066465.3
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr10_-_39901249 | 4.12 |
ENSMUST00000163705.3
|
Mfsd4b1
|
major facilitator superfamily domain containing 4B1 |
| chr12_-_28632514 | 4.06 |
ENSMUST00000110917.2
ENSMUST00000020965.14 |
Allc
|
allantoicase |
| chr5_-_144969564 | 4.05 |
ENSMUST00000071421.6
|
Gm4871
|
predicted gene 4871 |
| chr17_+_48400153 | 4.03 |
ENSMUST00000233043.2
|
1700067P10Rik
|
RIKEN cDNA 1700067P10 gene |
| chr14_+_16164930 | 3.99 |
ENSMUST00000112737.10
ENSMUST00000170923.9 |
Gm21103
|
predicted gene, 21103 |
| chr16_-_95993420 | 3.98 |
ENSMUST00000113804.8
ENSMUST00000054855.14 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chr5_+_144979796 | 3.96 |
ENSMUST00000031624.5
|
1700018F24Rik
|
RIKEN cDNA 1700018F24 gene |
| chrX_+_101952505 | 3.95 |
ENSMUST00000113602.2
|
1700011M02Rik
|
RIKEN cDNA 1700011M02 gene |
| chr2_-_129139125 | 3.95 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr19_+_5928649 | 3.94 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr5_+_146492946 | 3.93 |
ENSMUST00000179214.8
ENSMUST00000110595.3 |
Gm3415
|
predicted gene 3415 |
| chr4_+_116565819 | 3.93 |
ENSMUST00000106463.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr1_+_165957784 | 3.92 |
ENSMUST00000060833.14
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr5_+_146474457 | 3.92 |
ENSMUST00000199142.5
ENSMUST00000110597.4 ENSMUST00000110599.9 |
Gm3409
|
predicted gene 3409 |
| chr12_+_113112311 | 3.90 |
ENSMUST00000199089.5
|
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr11_-_99045894 | 3.86 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr2_+_148623929 | 3.84 |
ENSMUST00000028933.3
|
Cstdc1
|
cystatin domain containing 1 |
| chr17_+_25381414 | 3.83 |
ENSMUST00000073277.12
ENSMUST00000182621.8 |
Ccdc154
|
coiled-coil domain containing 154 |
| chr15_-_96917804 | 3.81 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr14_-_18538017 | 3.80 |
ENSMUST00000168306.8
|
Gm3029
|
predicted gene 3029 |
| chr17_-_35394971 | 3.77 |
ENSMUST00000173324.8
|
Aif1
|
allograft inflammatory factor 1 |
| chr14_-_17485779 | 3.76 |
ENSMUST00000171964.8
ENSMUST00000171636.8 |
Gm8297
|
predicted gene 8297 |
| chr10_+_128104525 | 3.75 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr5_-_24760427 | 3.74 |
ENSMUST00000198887.2
|
Iqca1l
|
IQ motif containing with AAA domain 1 like |
| chrX_-_101730578 | 3.74 |
ENSMUST00000122154.3
|
Gm3880
|
predicted pseudogene 3880 |
| chr2_+_152804405 | 3.71 |
ENSMUST00000099197.9
ENSMUST00000103155.10 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr13_-_99121070 | 3.70 |
ENSMUST00000054425.7
|
H2bl1
|
H2B.L histone variant 1 |
| chr5_+_42225303 | 3.69 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr14_-_17837363 | 3.67 |
ENSMUST00000170546.8
ENSMUST00000163738.8 |
Gm8159
|
predicted gene 8159 |
| chr11_+_105866030 | 3.65 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr7_+_75879603 | 3.64 |
ENSMUST00000156166.8
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr14_-_19652649 | 3.62 |
ENSMUST00000096121.12
|
Gm5458
|
predicted gene 5458 |
| chr1_+_175459559 | 3.62 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chrX_-_134985958 | 3.60 |
ENSMUST00000138878.2
ENSMUST00000080929.13 |
Nxf3
|
nuclear RNA export factor 3 |
| chr17_-_7620095 | 3.60 |
ENSMUST00000115747.3
|
Ttll2
|
tubulin tyrosine ligase-like family, member 2 |
| chr14_-_17338198 | 3.59 |
ENSMUST00000165713.8
ENSMUST00000112767.9 |
Gm8279
|
predicted gene 8279 |
| chr9_+_78137927 | 3.59 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr14_-_18835261 | 3.58 |
ENSMUST00000112802.9
|
Gm3012
|
predicted gene 3012 |
| chr14_-_19074680 | 3.58 |
ENSMUST00000112728.9
|
Gm10413
|
predicted gene 10413 |
| chr15_-_41733099 | 3.55 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr11_-_4390745 | 3.53 |
ENSMUST00000109948.8
|
Hormad2
|
HORMA domain containing 2 |
| chr15_+_75468473 | 3.52 |
ENSMUST00000189944.7
ENSMUST00000023243.11 |
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr8_+_21681630 | 3.49 |
ENSMUST00000098896.5
|
Defa31
|
defensin, alpha, 31 |
| chr2_-_24825066 | 3.45 |
ENSMUST00000132074.2
|
Arrdc1
|
arrestin domain containing 1 |
| chr14_-_21792938 | 3.45 |
ENSMUST00000120956.8
|
Dusp13
|
dual specificity phosphatase 13 |
| chr5_-_145656934 | 3.45 |
ENSMUST00000094111.6
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr7_+_43090206 | 3.43 |
ENSMUST00000040227.3
|
Cldnd2
|
claudin domain containing 2 |
| chr9_+_110248815 | 3.42 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr1_+_135060431 | 3.40 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr2_-_164621641 | 3.39 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr8_-_107792264 | 3.37 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_10135449 | 3.36 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr9_-_72892617 | 3.35 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr11_-_86999481 | 3.35 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr14_-_18304685 | 3.33 |
ENSMUST00000171866.8
ENSMUST00000167839.8 |
Gm3127
|
predicted gene 3127 |
| chr11_-_4391082 | 3.32 |
ENSMUST00000109949.8
ENSMUST00000130174.2 |
Hormad2
|
HORMA domain containing 2 |
| chr4_+_116565784 | 3.32 |
ENSMUST00000138305.8
ENSMUST00000125671.8 ENSMUST00000130828.8 |
Ccdc163
|
coiled-coil domain containing 163 |
| chr10_-_93375832 | 3.31 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr18_-_43610829 | 3.30 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr5_+_146428173 | 3.29 |
ENSMUST00000110611.8
ENSMUST00000198912.2 |
Gm6370
|
predicted gene 6370 |
| chr17_+_18108086 | 3.27 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr8_-_116434517 | 3.27 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr4_+_116565898 | 3.27 |
ENSMUST00000135499.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr17_-_13159204 | 3.25 |
ENSMUST00000043923.12
|
Acat3
|
acetyl-Coenzyme A acetyltransferase 3 |
| chr7_-_114162125 | 3.22 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr5_+_8010445 | 3.22 |
ENSMUST00000115421.3
|
Steap4
|
STEAP family member 4 |
| chr2_-_153286361 | 3.21 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like |
| chr11_+_58269862 | 3.20 |
ENSMUST00000013787.11
ENSMUST00000108826.3 |
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr1_+_165957909 | 3.20 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr4_+_150171822 | 3.20 |
ENSMUST00000094451.4
|
Gpr157
|
G protein-coupled receptor 157 |
| chr5_+_146450933 | 3.20 |
ENSMUST00000200228.5
ENSMUST00000036715.16 ENSMUST00000077133.7 |
Gm3402
|
predicted gene 3402 |
| chr14_+_15424803 | 3.19 |
ENSMUST00000169215.8
|
Gm3685
|
predicted gene 3685 |
| chr7_-_81216687 | 3.18 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr9_-_63056197 | 3.18 |
ENSMUST00000116613.9
|
Skor1
|
SKI family transcriptional corepressor 1 |
| chr2_+_153684901 | 3.17 |
ENSMUST00000175856.3
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr15_-_79626719 | 3.14 |
ENSMUST00000089311.11
ENSMUST00000046259.14 |
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr5_+_146418775 | 3.14 |
ENSMUST00000179032.3
|
Gm6408
|
predicted gene 6408 |
| chr14_+_15847355 | 3.13 |
ENSMUST00000167882.8
|
Gm3460
|
predicted gene 3460 |
| chr14_+_14874181 | 3.13 |
ENSMUST00000096148.11
|
Gm10338
|
predicted gene 10338 |
| chr6_+_29433247 | 3.12 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr12_+_101370932 | 3.12 |
ENSMUST00000055156.5
|
Catsperb
|
cation channel sperm associated auxiliary subunit beta |
| chr7_+_43751749 | 3.12 |
ENSMUST00000085455.6
|
Klk1b21
|
kallikrein 1-related peptidase b21 |
| chrX_-_154121454 | 3.12 |
ENSMUST00000026328.11
|
Prdx4
|
peroxiredoxin 4 |
| chr12_+_116239006 | 3.11 |
ENSMUST00000090195.5
|
Gm11027
|
predicted gene 11027 |
| chr7_-_126275529 | 3.11 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr17_+_24971952 | 3.10 |
ENSMUST00000044922.8
ENSMUST00000234202.2 |
Hs3st6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
| chr2_+_58644922 | 3.09 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr14_+_66043515 | 3.06 |
ENSMUST00000139644.2
|
Pbk
|
PDZ binding kinase |
| chr14_-_19281460 | 3.06 |
ENSMUST00000171150.8
ENSMUST00000100920.10 |
Gm5795
|
predicted gene 5795 |
| chr2_+_172314433 | 3.05 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr5_-_146107531 | 3.03 |
ENSMUST00000174320.2
|
Gm6309
|
predicted gene 6309 |
| chr5_+_146439209 | 3.03 |
ENSMUST00000110598.3
|
4930449I24Rik
|
RIKEN cDNA 4930449I24 gene |
| chr1_+_175459735 | 3.02 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_-_5610628 | 3.01 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr13_+_84370405 | 3.01 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr14_+_15104639 | 3.01 |
ENSMUST00000163826.2
|
Gm8374
|
predicted gene 8374 |
| chr19_+_12673147 | 3.00 |
ENSMUST00000025598.10
ENSMUST00000138545.8 ENSMUST00000154822.2 |
Keg1
|
kidney expressed gene 1 |
| chr2_+_164557219 | 2.99 |
ENSMUST00000103097.10
ENSMUST00000180193.2 |
Spint5
|
serine protease inhibitor, Kunitz type 5 |
| chr14_-_17987965 | 2.99 |
ENSMUST00000167902.2
ENSMUST00000169364.8 |
Gm3182
|
predicted gene 3182 |
| chr5_-_31854942 | 2.97 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr1_+_139429430 | 2.96 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr15_+_89452529 | 2.96 |
ENSMUST00000023295.3
ENSMUST00000230538.2 ENSMUST00000230978.2 |
Acr
|
acrosin prepropeptide |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 3.1 | 12.6 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 2.6 | 20.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.4 | 7.3 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 2.1 | 6.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 2.0 | 6.0 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 1.9 | 17.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.9 | 7.5 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 1.8 | 5.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.7 | 19.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 1.6 | 7.9 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 1.5 | 18.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.5 | 4.5 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 1.5 | 4.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 1.3 | 3.9 | GO:0097021 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.3 | 5.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.3 | 3.8 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 1.2 | 6.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 1.1 | 5.3 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 1.0 | 5.2 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.0 | 3.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.0 | 6.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.9 | 2.8 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.9 | 2.8 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.9 | 2.6 | GO:0046100 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 0.9 | 3.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.8 | 5.9 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.8 | 1.6 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.8 | 4.8 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.8 | 6.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.7 | 3.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 3.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.7 | 4.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.7 | 4.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.7 | 5.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.7 | 4.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.7 | 4.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.6 | 3.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.6 | 3.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.6 | 1.9 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.5 | 7.1 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.5 | 1.6 | GO:0009629 | response to gravity(GO:0009629) |
| 0.5 | 2.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.5 | 2.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.5 | 6.9 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.5 | 4.7 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.5 | 4.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.5 | 1.8 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.4 | 9.8 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.4 | 1.7 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.4 | 2.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 1.2 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.4 | 3.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.4 | 4.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.4 | 2.9 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.4 | 3.6 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.4 | 4.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 2.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 2.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.3 | 2.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 5.0 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.3 | 3.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 3.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 2.2 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.3 | 0.9 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.3 | 2.1 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.3 | 1.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 4.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 1.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.3 | 5.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 3.9 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.2 | 1.9 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 6.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 0.9 | GO:0061739 | aggrephagy(GO:0035973) protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.2 | 1.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 4.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.3 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.2 | 2.7 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 1.6 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 | 1.6 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 30.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 3.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.2 | 5.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 1.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 2.3 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.2 | 3.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.2 | 1.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 1.7 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.2 | 3.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 4.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 4.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 0.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 2.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 3.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.4 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 16.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 7.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 2.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.6 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 4.0 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.1 | 1.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 3.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 5.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 4.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 3.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.3 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 26.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 3.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 2.2 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 1.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 3.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 3.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 2.9 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 2.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 3.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 4.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 3.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 1.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 1.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 3.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 1.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 2.2 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 2.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 3.1 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 1.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 7.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 2.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.7 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.9 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.9 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.0 | 1.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 3.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 2.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 1.5 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 2.9 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.3 | 11.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.2 | 3.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.9 | 4.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.7 | 3.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.7 | 29.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.7 | 5.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 6.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.7 | 5.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 2.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.5 | 2.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.5 | 5.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.5 | 8.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 1.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 1.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.4 | 2.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 2.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.3 | 2.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 2.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 0.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 5.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 8.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 4.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 9.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 3.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 2.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 2.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 0.6 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.2 | 6.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.7 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.2 | 3.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 3.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 20.5 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.8 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 2.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 2.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 3.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 13.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 7.8 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 3.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 4.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 14.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 2.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 12.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 7.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 7.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 66.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 5.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 1.8 | 5.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 1.7 | 1.7 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 1.7 | 6.9 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 1.7 | 13.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 1.6 | 11.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.6 | 4.8 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.5 | 4.6 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 1.3 | 7.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.2 | 6.0 | GO:0035478 | chylomicron binding(GO:0035478) |
| 1.1 | 7.9 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 1.1 | 5.5 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.1 | 12.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 1.0 | 7.7 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.9 | 4.6 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.9 | 5.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.8 | 6.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.8 | 14.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.8 | 3.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.7 | 2.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.6 | 4.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.6 | 3.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.6 | 3.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.6 | 1.8 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.6 | 2.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.6 | 14.9 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.5 | 1.6 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.5 | 1.6 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.5 | 4.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.5 | 3.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.5 | 1.5 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.5 | 1.4 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.5 | 3.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.4 | 18.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 7.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.4 | 5.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 4.3 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 6.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 2.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.4 | 3.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 4.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 3.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.4 | 3.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 6.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 7.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.3 | 0.9 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.3 | 16.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.3 | 3.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 8.6 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 1.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 4.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 0.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 1.4 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.3 | 5.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 1.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 9.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 1.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.7 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.2 | 1.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 1.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 27.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.9 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 3.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.6 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 2.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.6 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.2 | 1.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 1.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 4.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 3.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 5.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 3.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 5.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 19.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 4.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 2.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 4.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 19.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.1 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 2.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 10.1 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 2.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 6.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 7.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 3.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 9.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.4 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 3.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 3.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 1.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 5.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 3.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 5.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 5.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 17.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 7.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 6.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 7.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 15.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 5.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 4.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 17.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 8.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.6 | 4.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 9.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.4 | 5.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 4.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 4.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 5.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 7.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 4.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 5.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 3.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 5.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 7.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 10.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 7.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 8.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 7.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 11.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 4.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 4.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 8.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 10.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 7.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.6 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 3.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |