Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Arid5a
Z-value: 0.51
Transcription factors associated with Arid5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5a
|
ENSMUSG00000037447.17 | Arid5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid5a | mm39_v1_chr1_+_36346824_36346850 | -0.35 | 2.4e-03 | Click! |
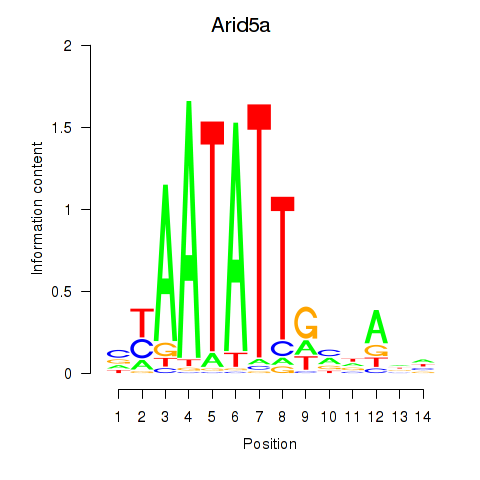
Activity profile of Arid5a motif
Sorted Z-values of Arid5a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_58645189 | 6.35 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr7_-_105249308 | 5.37 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr2_+_58644922 | 4.84 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr5_-_87074380 | 4.80 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr2_+_61876956 | 3.29 |
ENSMUST00000112480.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_61876923 | 3.11 |
ENSMUST00000054484.15
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_61876806 | 2.95 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr15_+_41573995 | 2.55 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr2_-_79959178 | 2.05 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_+_75982890 | 1.72 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr1_+_133965228 | 1.63 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr15_-_55421144 | 1.55 |
ENSMUST00000172387.8
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr2_+_105505823 | 1.55 |
ENSMUST00000167211.9
ENSMUST00000111083.10 |
Pax6
|
paired box 6 |
| chr2_-_79959802 | 1.24 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr18_+_37637317 | 1.09 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr4_+_43384320 | 0.98 |
ENSMUST00000136360.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr14_+_58313964 | 0.91 |
ENSMUST00000166770.2
|
Fgf9
|
fibroblast growth factor 9 |
| chr10_-_115197775 | 0.79 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr10_-_115198093 | 0.78 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr9_+_20148415 | 0.76 |
ENSMUST00000086474.6
|
Olfr872
|
olfactory receptor 872 |
| chrM_+_7758 | 0.71 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chrM_+_7779 | 0.67 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr16_-_58583539 | 0.67 |
ENSMUST00000214139.2
|
Olfr172
|
olfactory receptor 172 |
| chr3_-_146487102 | 0.56 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr1_-_171885140 | 0.56 |
ENSMUST00000059794.4
|
Nhlh1
|
nescient helix loop helix 1 |
| chr3_+_122298308 | 0.37 |
ENSMUST00000199358.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr10_+_52267702 | 0.35 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr11_-_58425662 | 0.28 |
ENSMUST00000213188.3
|
Olfr330
|
olfactory receptor 330 |
| chr10_+_69369854 | 0.20 |
ENSMUST00000182557.8
|
Ank3
|
ankyrin 3, epithelial |
| chr10_-_129530155 | 0.20 |
ENSMUST00000204641.3
|
Olfr803
|
olfactory receptor 803 |
| chrM_+_8603 | 0.15 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr10_+_129298547 | 0.11 |
ENSMUST00000077836.3
|
Olfr787
|
olfactory receptor 787 |
| chr17_-_37627945 | 0.07 |
ENSMUST00000217590.2
|
Olfr102
|
olfactory receptor 102 |
| chrX_-_133012600 | 0.04 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.8 | 5.4 | GO:0015886 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.4 | 1.5 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.3 | 9.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 3.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 11.2 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.1 | 0.9 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 1.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.0 | 11.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.9 | 5.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 9.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 3.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 4.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.5 | GO:0070410 | AT DNA binding(GO:0003680) co-SMAD binding(GO:0070410) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 5.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |