Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Arid5b
Z-value: 0.76
Transcription factors associated with Arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5b
|
ENSMUSG00000019947.11 | Arid5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid5b | mm39_v1_chr10_-_68114543_68114570 | 0.02 | 8.7e-01 | Click! |
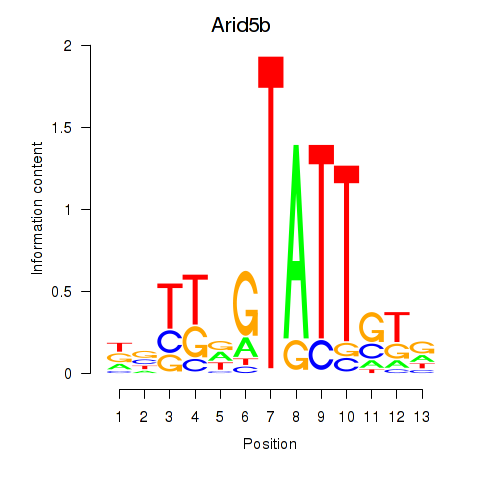
Activity profile of Arid5b motif
Sorted Z-values of Arid5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_92535705 | 5.71 |
ENSMUST00000138687.2
ENSMUST00000124509.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr5_+_92535532 | 4.99 |
ENSMUST00000145072.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr12_+_103524690 | 4.58 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr8_-_13304154 | 4.57 |
ENSMUST00000204916.3
ENSMUST00000033825.11 |
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr18_+_23548534 | 3.82 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr17_+_44112187 | 3.71 |
ENSMUST00000228972.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr8_-_13304068 | 3.55 |
ENSMUST00000168498.8
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr8_-_13304096 | 3.38 |
ENSMUST00000171619.2
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr14_+_70077841 | 3.29 |
ENSMUST00000022678.5
|
Pebp4
|
phosphatidylethanolamine binding protein 4 |
| chr14_-_75829389 | 3.13 |
ENSMUST00000165569.3
ENSMUST00000035243.5 |
Cby2
|
chibby family member 2 |
| chr1_+_190769010 | 3.05 |
ENSMUST00000077889.8
|
Spata45
|
spermatogenesis associated 45 |
| chr11_+_70506716 | 3.04 |
ENSMUST00000144960.2
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr18_+_23548192 | 2.94 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr18_+_23548455 | 2.80 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr7_-_44624165 | 2.59 |
ENSMUST00000212836.2
ENSMUST00000212255.2 ENSMUST00000063761.8 |
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr11_+_70506674 | 2.57 |
ENSMUST00000180052.8
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr15_+_41615084 | 2.55 |
ENSMUST00000229511.2
ENSMUST00000229836.2 |
Oxr1
|
oxidation resistance 1 |
| chr3_-_95789505 | 2.50 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr7_-_67294943 | 2.29 |
ENSMUST00000190276.7
ENSMUST00000032775.12 ENSMUST00000053950.10 ENSMUST00000189836.2 |
Lrrc28
|
leucine rich repeat containing 28 |
| chr6_-_23650205 | 2.25 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr9_-_8134295 | 2.21 |
ENSMUST00000037397.8
|
Cep126
|
centrosomal protein 126 |
| chr1_+_163607143 | 2.17 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr5_-_37146266 | 2.13 |
ENSMUST00000166339.8
|
Wfs1
|
wolframin ER transmembrane glycoprotein |
| chr4_-_154752320 | 2.12 |
ENSMUST00000060062.5
|
Actrt2
|
actin-related protein T2 |
| chr2_-_110591909 | 2.12 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr6_-_23650297 | 2.05 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr7_-_101570393 | 2.04 |
ENSMUST00000106965.8
ENSMUST00000106968.8 ENSMUST00000106967.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr7_-_127185512 | 1.97 |
ENSMUST00000205839.2
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr12_-_11485639 | 1.96 |
ENSMUST00000220506.2
|
Vsnl1
|
visinin-like 1 |
| chr12_+_113507528 | 1.96 |
ENSMUST00000053086.3
|
Adam6a
|
a disintegrin and metallopeptidase domain 6A |
| chr1_+_100025486 | 1.95 |
ENSMUST00000188735.2
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr5_+_31454787 | 1.88 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr15_+_51903503 | 1.86 |
ENSMUST00000090025.5
|
Aard
|
alanine and arginine rich domain containing protein |
| chr16_-_38649107 | 1.86 |
ENSMUST00000122078.3
|
Tex55
|
testis expressed 55 |
| chr8_-_43594523 | 1.68 |
ENSMUST00000059692.4
|
Triml1
|
tripartite motif family-like 1 |
| chr4_-_118347249 | 1.64 |
ENSMUST00000047421.6
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr17_+_69071546 | 1.61 |
ENSMUST00000233625.2
|
L3mbtl4
|
L3MBTL4 histone methyl-lysine binding protein |
| chr7_-_12819142 | 1.61 |
ENSMUST00000094829.2
|
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr18_+_37575553 | 1.61 |
ENSMUST00000056915.3
|
Pcdhb13
|
protocadherin beta 13 |
| chr7_+_108266625 | 1.60 |
ENSMUST00000076289.2
|
Olfr510
|
olfactory receptor 510 |
| chr2_-_84509172 | 1.57 |
ENSMUST00000111665.8
|
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr9_-_58065800 | 1.56 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr6_+_120643323 | 1.54 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr3_+_32583681 | 1.53 |
ENSMUST00000147350.8
|
Mfn1
|
mitofusin 1 |
| chr1_+_174000304 | 1.49 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr2_-_84508385 | 1.43 |
ENSMUST00000189772.2
ENSMUST00000053664.9 ENSMUST00000111664.8 |
Gm28635
Tmx2
|
predicted gene 28635 thioredoxin-related transmembrane protein 2 |
| chrX_+_139357362 | 1.41 |
ENSMUST00000033809.4
|
Prps1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr17_+_32671689 | 1.38 |
ENSMUST00000237491.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr13_-_96269076 | 1.37 |
ENSMUST00000161263.8
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chrX_-_156275231 | 1.36 |
ENSMUST00000112529.8
|
Sms
|
spermine synthase |
| chr4_+_116030561 | 1.35 |
ENSMUST00000106492.3
ENSMUST00000216692.2 |
1700042G07Rik
Gm49337
|
RIKEN cDNA 1700042G07 gene predicted gene, 49337 |
| chrX_+_81992467 | 1.31 |
ENSMUST00000114000.8
|
Dmd
|
dystrophin, muscular dystrophy |
| chr7_+_29008840 | 1.28 |
ENSMUST00000137848.2
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr17_+_71326542 | 1.27 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr9_-_45895635 | 1.22 |
ENSMUST00000215427.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr7_+_30681287 | 1.20 |
ENSMUST00000128384.3
|
Fam187b
|
family with sequence similarity 187, member B |
| chr17_+_71326510 | 1.17 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr2_-_160155536 | 1.17 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr19_+_22670134 | 1.16 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr1_+_31261889 | 1.16 |
ENSMUST00000027230.3
|
Dnaaf6
|
dynein axonemal assembly factor 6 |
| chr2_-_111320501 | 1.08 |
ENSMUST00000099616.2
|
Olfr1290
|
olfactory receptor 1290 |
| chrX_-_111316476 | 1.07 |
ENSMUST00000026601.3
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chrM_-_14061 | 1.03 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr10_+_57660948 | 0.98 |
ENSMUST00000020024.12
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr9_-_101076198 | 0.98 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_+_111136546 | 0.95 |
ENSMUST00000090329.2
|
Olfr1279
|
olfactory receptor 1279 |
| chr6_-_68857658 | 0.88 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr4_-_148021159 | 0.87 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr11_+_11414256 | 0.84 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr16_+_32315090 | 0.83 |
ENSMUST00000231510.2
|
Zdhhc19
|
zinc finger, DHHC domain containing 19 |
| chr12_-_91712783 | 0.81 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr6_+_68451563 | 0.81 |
ENSMUST00000200406.2
|
Igkv20-101-2
|
immunoglobulin kappa chain variable 20-101-2 |
| chr11_+_74255497 | 0.80 |
ENSMUST00000077794.4
|
Olfr412
|
olfactory receptor 412 |
| chr11_-_100084072 | 0.80 |
ENSMUST00000059707.3
|
Krt9
|
keratin 9 |
| chr19_-_44017637 | 0.80 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr10_-_108846816 | 0.80 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr11_-_32217547 | 0.78 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr3_-_19319123 | 0.77 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr11_+_73131136 | 0.72 |
ENSMUST00000138853.3
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr2_-_77349909 | 0.71 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr2_-_151586063 | 0.70 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr13_+_112797273 | 0.70 |
ENSMUST00000052514.6
|
Slc38a9
|
solute carrier family 38, member 9 |
| chr7_+_107166925 | 0.67 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr17_-_35937526 | 0.66 |
ENSMUST00000164502.3
|
Muc21
|
mucin 21 |
| chr8_+_106620174 | 0.66 |
ENSMUST00000060167.6
ENSMUST00000118920.2 |
Nrn1l
|
neuritin 1-like |
| chr3_+_59637203 | 0.65 |
ENSMUST00000168156.3
|
Aadacl2fm2
|
AADACL2 family member 2 |
| chr7_+_107166653 | 0.64 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr4_-_150993886 | 0.62 |
ENSMUST00000128075.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr19_-_7688628 | 0.61 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr9_+_57913694 | 0.60 |
ENSMUST00000188116.7
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr6_-_57821483 | 0.58 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr2_+_84508734 | 0.58 |
ENSMUST00000102645.4
|
Med19
|
mediator complex subunit 19 |
| chr18_+_36661198 | 0.58 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr3_-_20296337 | 0.57 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr14_+_26414422 | 0.56 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr7_+_114342929 | 0.55 |
ENSMUST00000161800.2
|
Insc
|
INSC spindle orientation adaptor protein |
| chr9_+_104930438 | 0.53 |
ENSMUST00000149243.8
ENSMUST00000035177.15 ENSMUST00000214036.2 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chrM_+_8603 | 0.53 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr1_+_180158035 | 0.52 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr8_-_62576140 | 0.52 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr18_+_37864045 | 0.51 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr9_+_24194729 | 0.51 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr15_-_55411560 | 0.50 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr15_-_13173736 | 0.50 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr17_+_36857967 | 0.48 |
ENSMUST00000041964.7
|
H2-M11
|
histocompatibility 2, M region locus 11 |
| chr14_-_26256025 | 0.48 |
ENSMUST00000139075.8
ENSMUST00000102956.8 |
Slmap
|
sarcolemma associated protein |
| chr11_-_109188892 | 0.48 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr17_-_37404764 | 0.47 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr7_-_103113358 | 0.46 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr4_-_129556234 | 0.43 |
ENSMUST00000003828.11
|
Kpna6
|
karyopherin (importin) alpha 6 |
| chr7_+_33794856 | 0.42 |
ENSMUST00000108083.2
|
Scgb1b30
|
secretoglobin, family 1B, member 30 |
| chr18_+_65022035 | 0.41 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr16_+_57173632 | 0.38 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr13_-_96607722 | 0.37 |
ENSMUST00000055607.13
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr9_-_39237341 | 0.36 |
ENSMUST00000216132.2
|
Olfr948
|
olfactory receptor 948 |
| chr3_+_59768472 | 0.36 |
ENSMUST00000179799.2
|
Aadacl2fm3
|
AADACL2 family member 3 |
| chr7_+_107679062 | 0.35 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr2_-_34716083 | 0.35 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr3_-_89905547 | 0.34 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr17_+_32671719 | 0.32 |
ENSMUST00000236307.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr6_-_40521911 | 0.32 |
ENSMUST00000089490.3
|
Olfr461
|
olfactory receptor 461 |
| chr9_-_45896075 | 0.31 |
ENSMUST00000217636.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr14_+_52804637 | 0.27 |
ENSMUST00000196939.2
ENSMUST00000103568.3 |
Trav2
|
T cell receptor alpha variable 2 |
| chr2_+_111084861 | 0.27 |
ENSMUST00000218065.2
|
Olfr1276
|
olfactory receptor 1276 |
| chr19_+_13339600 | 0.26 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chr10_-_130002635 | 0.26 |
ENSMUST00000216530.2
|
Olfr825
|
olfactory receptor 825 |
| chr2_+_111329683 | 0.25 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr6_+_121323577 | 0.25 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr7_+_104199340 | 0.23 |
ENSMUST00000216904.2
|
Olfr651
|
olfactory receptor 651 |
| chr7_+_31075005 | 0.22 |
ENSMUST00000179481.2
|
Scgb1b3
|
secretoglobin, family 1B, member 3 |
| chr18_+_44249254 | 0.21 |
ENSMUST00000212114.2
|
Gm37797
|
predicted gene, 37797 |
| chr6_+_24748324 | 0.21 |
ENSMUST00000031691.3
|
Hyal4
|
hyaluronoglucosaminidase 4 |
| chr11_+_50917831 | 0.20 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr3_-_89905927 | 0.20 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr10_+_102376109 | 0.20 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr18_-_70409277 | 0.18 |
ENSMUST00000239144.2
|
Gm36255
|
predicted gene, 36255 |
| chr5_+_42225303 | 0.17 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr17_-_37543259 | 0.17 |
ENSMUST00000207414.3
|
Olfr97
|
olfactory receptor 97 |
| chr12_-_24730901 | 0.16 |
ENSMUST00000156453.10
|
Cys1
|
cystin 1 |
| chr19_+_12972378 | 0.16 |
ENSMUST00000207997.3
|
Olfr1451
|
olfactory receptor 1451 |
| chr1_+_88154727 | 0.15 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chrX_+_8929254 | 0.15 |
ENSMUST00000097029.4
|
Fthl17f
|
ferritin, heavy polypeptide-like 17, member F |
| chr11_-_99276868 | 0.15 |
ENSMUST00000211768.2
|
Krt10
|
keratin 10 |
| chr16_-_26345493 | 0.14 |
ENSMUST00000165687.3
|
Tmem207
|
transmembrane protein 207 |
| chr6_-_133014064 | 0.14 |
ENSMUST00000032317.4
|
Tas2r103
|
taste receptor, type 2, member 103 |
| chr13_-_22805305 | 0.13 |
ENSMUST00000187140.3
|
Vmn1r206
|
vomeronasal 1 receptor 206 |
| chr9_-_45896110 | 0.12 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr16_-_17540805 | 0.11 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr19_-_12147438 | 0.11 |
ENSMUST00000207679.3
ENSMUST00000219261.2 |
Olfr1555-ps1
|
olfactory receptor 1555, pseudogene 1 |
| chr14_+_53284744 | 0.11 |
ENSMUST00000103606.2
|
Trav16d-dv11
|
T cell receptor alpha variable 16D-DV11 |
| chr17_+_37670473 | 0.08 |
ENSMUST00000178766.3
ENSMUST00000215398.2 |
Olfr104-ps
|
olfactory receptor 104, pseudogene |
| chr19_-_12209960 | 0.07 |
ENSMUST00000207710.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr1_+_31215482 | 0.03 |
ENSMUST00000062560.14
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr6_+_106095726 | 0.00 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr2_+_111198936 | 0.00 |
ENSMUST00000090328.5
|
Olfr1283
|
olfactory receptor 1283 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.1 | 4.6 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.7 | 2.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.5 | 1.4 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.5 | 1.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 1.3 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.3 | 1.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 | 1.9 | GO:0009750 | response to fructose(GO:0009750) |
| 0.3 | 2.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.2 | 10.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 1.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 0.7 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.2 | 0.9 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 2.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.6 | GO:0018323 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) regulation of cellular amino acid biosynthetic process(GO:2000282) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.2 | 3.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 2.6 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 2.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.0 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.5 | GO:2000293 | defecation(GO:0030421) negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 1.5 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.5 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.6 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.1 | 2.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 2.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 4.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.6 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 2.0 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 1.6 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 3.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 2.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 1.7 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.7 | 2.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.5 | 1.5 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 1.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 1.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.5 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 2.4 | GO:0032982 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 0.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 5.6 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 2.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 11.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.8 | 10.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 2.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 3.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 1.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 1.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.7 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 1.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 0.9 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 0.6 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.2 | 0.6 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.1 | 2.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 2.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 9.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 4.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 2.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.0 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |