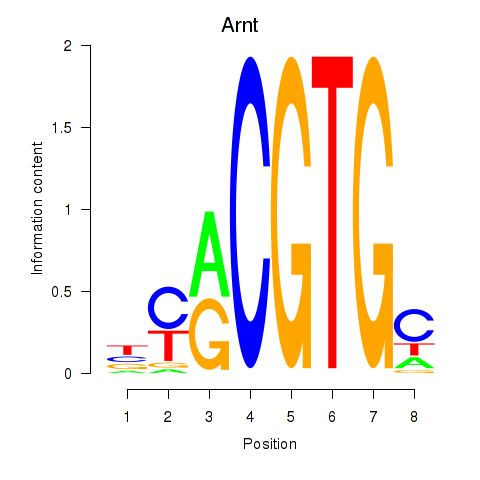
|
chr6_-_52168675
Show fit
|
6.47 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4
|
|
chr10_+_97528915
Show fit
|
5.71 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1
|
|
chr3_+_130904000
Show fit
|
4.85 |
ENSMUST00000029611.14
ENSMUST00000106341.9
ENSMUST00000066849.13
|
Lef1
|
lymphoid enhancer binding factor 1
|
|
chr2_+_122065230
Show fit
|
3.74 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase
|
|
chr2_-_37593856
Show fit
|
3.51 |
ENSMUST00000155237.2
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr17_-_5468938
Show fit
|
3.17 |
ENSMUST00000189788.2
|
Ldhal6b
|
lactate dehydrogenase A-like 6B
|
|
chr6_+_64706101
Show fit
|
3.11 |
ENSMUST00000101351.6
|
Atoh1
|
atonal bHLH transcription factor 1
|
|
chr17_+_27775613
Show fit
|
2.98 |
ENSMUST00000231780.2
ENSMUST00000232253.2
ENSMUST00000232552.2
ENSMUST00000117600.9
|
Hmga1
|
high mobility group AT-hook 1
|
|
chr9_-_62888156
Show fit
|
2.93 |
ENSMUST00000098651.6
ENSMUST00000214830.2
|
Pias1
|
protein inhibitor of activated STAT 1
|
|
chr19_+_41970148
Show fit
|
2.90 |
ENSMUST00000026170.3
|
Ubtd1
|
ubiquitin domain containing 1
|
|
chr17_+_27775637
Show fit
|
2.88 |
ENSMUST00000117254.9
ENSMUST00000231243.2
ENSMUST00000231358.2
ENSMUST00000118570.2
ENSMUST00000231796.2
|
Hmga1
|
high mobility group AT-hook 1
|
|
chr6_-_99703344
Show fit
|
2.84 |
ENSMUST00000008273.8
ENSMUST00000101120.11
ENSMUST00000203738.2
|
Prok2
|
prokineticin 2
|
|
chr13_+_108452866
Show fit
|
2.81 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B
|
|
chr10_+_80100812
Show fit
|
2.68 |
ENSMUST00000105362.8
ENSMUST00000105361.10
|
Dazap1
|
DAZ associated protein 1
|
|
chr15_-_36609208
Show fit
|
2.65 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr4_-_34882917
Show fit
|
2.60 |
ENSMUST00000098163.9
ENSMUST00000047950.6
|
Zfp292
|
zinc finger protein 292
|
|
chr3_-_94693780
Show fit
|
2.59 |
ENSMUST00000107273.9
ENSMUST00000238849.2
|
Cgn
|
cingulin
|
|
chr5_-_137784912
Show fit
|
2.49 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme
|
|
chr14_+_69409251
Show fit
|
2.45 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6
|
|
chr10_-_4337435
Show fit
|
2.45 |
ENSMUST00000100077.5
|
Zbtb2
|
zinc finger and BTB domain containing 2
|
|
chr1_+_87254729
Show fit
|
2.43 |
ENSMUST00000172794.8
ENSMUST00000164992.9
ENSMUST00000173173.8
|
Gigyf2
|
GRB10 interacting GYF protein 2
|
|
chr8_-_46747629
Show fit
|
2.42 |
ENSMUST00000058636.9
|
Helt
|
helt bHLH transcription factor
|
|
chr17_+_27775471
Show fit
|
2.41 |
ENSMUST00000118599.9
ENSMUST00000232265.2
ENSMUST00000232013.2
ENSMUST00000114888.11
ENSMUST00000231874.2
ENSMUST00000119486.9
ENSMUST00000231825.2
ENSMUST00000231866.2
|
Hmga1
|
high mobility group AT-hook 1
|
|
chr3_+_89680867
Show fit
|
2.33 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr10_-_80223475
Show fit
|
2.32 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D
|
|
chr10_-_79602764
Show fit
|
2.30 |
ENSMUST00000047203.9
|
Rnf126
|
ring finger protein 126
|
|
chr5_-_137784943
Show fit
|
2.20 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme
|
|
chr2_+_119727689
Show fit
|
2.20 |
ENSMUST00000046717.13
ENSMUST00000079934.12
ENSMUST00000110774.8
ENSMUST00000110773.9
ENSMUST00000156510.2
|
Mga
|
MAX gene associated
|
|
chr3_-_94693740
Show fit
|
2.19 |
ENSMUST00000153263.9
ENSMUST00000107272.7
ENSMUST00000155485.4
|
Cgn
|
cingulin
|
|
chr13_+_108452930
Show fit
|
2.18 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B
|
|
chr1_-_178165223
Show fit
|
2.18 |
ENSMUST00000037748.9
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr10_-_18891095
Show fit
|
2.08 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_-_6327884
Show fit
|
2.08 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene
|
|
chr5_+_28370687
Show fit
|
2.07 |
ENSMUST00000036177.9
|
En2
|
engrailed 2
|
|
chr17_+_36172235
Show fit
|
2.02 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein)
|
|
chr13_-_112788829
Show fit
|
2.01 |
ENSMUST00000075748.7
|
Ddx4
|
DEAD box helicase 4
|
|
chr17_+_36172210
Show fit
|
1.98 |
ENSMUST00000074259.15
ENSMUST00000174873.2
|
Nrm
|
nurim (nuclear envelope membrane protein)
|
|
chr9_-_103243039
Show fit
|
1.97 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3
|
|
chr2_+_84669739
Show fit
|
1.95 |
ENSMUST00000146816.8
ENSMUST00000028469.14
|
Slc43a1
|
solute carrier family 43, member 1
|
|
chr19_+_41818409
Show fit
|
1.94 |
ENSMUST00000087155.5
|
Frat1
|
frequently rearranged in advanced T cell lymphomas
|
|
chr13_-_112788890
Show fit
|
1.93 |
ENSMUST00000099166.10
|
Ddx4
|
DEAD box helicase 4
|
|
chr11_+_93776965
Show fit
|
1.93 |
ENSMUST00000063718.11
ENSMUST00000107854.9
|
Mbtd1
|
mbt domain containing 1
|
|
chr16_+_35590745
Show fit
|
1.90 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1
|
|
chr11_+_93776650
Show fit
|
1.87 |
ENSMUST00000107853.8
ENSMUST00000107850.8
|
Mbtd1
|
mbt domain containing 1
|
|
chr15_+_80556023
Show fit
|
1.85 |
ENSMUST00000023044.7
|
Fam83f
|
family with sequence similarity 83, member F
|
|
chr11_-_70545424
Show fit
|
1.83 |
ENSMUST00000108549.2
|
Pfn1
|
profilin 1
|
|
chr1_+_87254719
Show fit
|
1.79 |
ENSMUST00000027475.15
|
Gigyf2
|
GRB10 interacting GYF protein 2
|
|
chr2_+_127967951
Show fit
|
1.76 |
ENSMUST00000089634.12
ENSMUST00000019281.14
ENSMUST00000110341.9
ENSMUST00000103211.8
ENSMUST00000103210.2
|
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr8_+_80366247
Show fit
|
1.72 |
ENSMUST00000173078.8
ENSMUST00000173286.8
|
Otud4
|
OTU domain containing 4
|
|
chr17_+_56347424
Show fit
|
1.71 |
ENSMUST00000002914.10
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr19_+_6952319
Show fit
|
1.71 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B
|
|
chr6_-_100264439
Show fit
|
1.71 |
ENSMUST00000101118.4
|
Rybp
|
RING1 and YY1 binding protein
|
|
chr9_-_103242737
Show fit
|
1.68 |
ENSMUST00000072249.13
ENSMUST00000189896.2
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3
|
|
chr19_-_7218363
Show fit
|
1.67 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit
|
|
chr5_-_148988413
Show fit
|
1.66 |
ENSMUST00000093196.11
|
Hmgb1
|
high mobility group box 1
|
|
chr19_+_41471067
Show fit
|
1.66 |
ENSMUST00000067795.13
|
Lcor
|
ligand dependent nuclear receptor corepressor
|
|
chr16_+_4867876
Show fit
|
1.64 |
ENSMUST00000230703.2
ENSMUST00000052449.6
|
Ubn1
|
ubinuclein 1
|
|
chr1_-_75156993
Show fit
|
1.63 |
ENSMUST00000027396.15
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr18_+_14916295
Show fit
|
1.62 |
ENSMUST00000234789.2
ENSMUST00000169862.2
|
Taf4b
|
TATA-box binding protein associated factor 4b
|
|
chr8_+_56747613
Show fit
|
1.60 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD)
|
|
chr10_+_69048464
Show fit
|
1.58 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1
|
|
chr12_+_72488625
Show fit
|
1.55 |
ENSMUST00000161284.3
ENSMUST00000162159.8
|
Lrrc9
|
leucine rich repeat containing 9
|
|
chr9_-_110483210
Show fit
|
1.54 |
ENSMUST00000196488.5
ENSMUST00000133191.8
ENSMUST00000167320.8
|
Nbeal2
|
neurobeachin-like 2
|
|
chr11_+_118913788
Show fit
|
1.54 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2
|
|
chr11_+_61575245
Show fit
|
1.52 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G
|
|
chr19_-_7218512
Show fit
|
1.52 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit
|
|
chr7_-_26865838
Show fit
|
1.51 |
ENSMUST00000108382.2
|
Egln2
|
egl-9 family hypoxia-inducible factor 2
|
|
chr8_+_91635192
Show fit
|
1.50 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9
|
|
chr3_-_88204286
Show fit
|
1.49 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone
|
|
chr15_+_99476935
Show fit
|
1.48 |
ENSMUST00000023752.6
|
Aqp2
|
aquaporin 2
|
|
chr17_+_24971952
Show fit
|
1.46 |
ENSMUST00000044922.8
ENSMUST00000234202.2
|
Hs3st6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6
|
|
chr1_-_74544946
Show fit
|
1.46 |
ENSMUST00000044260.11
ENSMUST00000186282.7
|
Usp37
|
ubiquitin specific peptidase 37
|
|
chr10_+_69048506
Show fit
|
1.44 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1
|
|
chr15_-_36609812
Show fit
|
1.44 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr3_-_89959917
Show fit
|
1.44 |
ENSMUST00000197903.5
|
Ubap2l
|
ubiquitin-associated protein 2-like
|
|
chr7_+_15795735
Show fit
|
1.44 |
ENSMUST00000209369.2
|
Zfp541
|
zinc finger protein 541
|
|
chr13_-_104246084
Show fit
|
1.44 |
ENSMUST00000224945.2
ENSMUST00000109315.5
|
Nln
|
neurolysin (metallopeptidase M3 family)
|
|
chr2_-_38816229
Show fit
|
1.43 |
ENSMUST00000076275.11
ENSMUST00000142130.2
|
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr3_-_89959770
Show fit
|
1.43 |
ENSMUST00000029553.16
ENSMUST00000195995.5
ENSMUST00000064639.15
ENSMUST00000199834.5
|
Ubap2l
|
ubiquitin-associated protein 2-like
|
|
chr17_-_65920481
Show fit
|
1.41 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A
|
|
chr1_+_84817547
Show fit
|
1.40 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36
|
|
chr10_+_127512933
Show fit
|
1.39 |
ENSMUST00000118612.8
ENSMUST00000048099.5
|
Nemp1
|
nuclear envelope integral membrane protein 1
|
|
chr7_+_46445512
Show fit
|
1.39 |
ENSMUST00000006774.11
ENSMUST00000165031.8
|
Gtf2h1
|
general transcription factor II H, polypeptide 1
|
|
chr6_-_52203146
Show fit
|
1.39 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9
|
|
chr1_-_181039509
Show fit
|
1.39 |
ENSMUST00000162819.9
ENSMUST00000237749.2
|
Wdr26
|
WD repeat domain 26
|
|
chr3_-_89959739
Show fit
|
1.37 |
ENSMUST00000199929.2
ENSMUST00000090908.11
ENSMUST00000198322.5
ENSMUST00000196843.5
|
Ubap2l
|
ubiquitin-associated protein 2-like
|
|
chr11_+_69805005
Show fit
|
1.37 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2
|
|
chr2_-_163261439
Show fit
|
1.34 |
ENSMUST00000046908.10
|
Oser1
|
oxidative stress responsive serine rich 1
|
|
chr8_+_84724130
Show fit
|
1.33 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1
|
|
chr12_-_59266511
Show fit
|
1.33 |
ENSMUST00000043204.8
|
Fbxo33
|
F-box protein 33
|
|
chr12_+_80837284
Show fit
|
1.32 |
ENSMUST00000220238.2
ENSMUST00000068519.7
|
Susd6
|
sushi domain containing 6
|
|
chr9_-_13738204
Show fit
|
1.28 |
ENSMUST00000147115.8
|
Cep57
|
centrosomal protein 57
|
|
chr10_-_62322551
Show fit
|
1.28 |
ENSMUST00000105447.11
|
Vps26a
|
VPS26 retromer complex component A
|
|
chr1_-_75119277
Show fit
|
1.28 |
ENSMUST00000168720.8
ENSMUST00000041213.12
ENSMUST00000189809.2
|
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr3_+_137570334
Show fit
|
1.27 |
ENSMUST00000174561.8
ENSMUST00000173790.8
|
H2az1
|
H2A.Z variant histone 1
|
|
chrX_+_7439839
Show fit
|
1.27 |
ENSMUST00000144719.9
ENSMUST00000234896.2
|
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA
forkhead box P3
|
|
chr1_-_75195127
Show fit
|
1.26 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A
|
|
chr14_-_31552335
Show fit
|
1.25 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28
|
|
chr11_+_98828495
Show fit
|
1.24 |
ENSMUST00000107475.9
ENSMUST00000068133.10
|
Rara
|
retinoic acid receptor, alpha
|
|
chr13_-_38178059
Show fit
|
1.23 |
ENSMUST00000225319.2
ENSMUST00000225246.2
ENSMUST00000021864.8
|
Ssr1
|
signal sequence receptor, alpha
|
|
chr1_+_74545203
Show fit
|
1.23 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9
|
|
chr19_+_41471395
Show fit
|
1.23 |
ENSMUST00000237208.2
ENSMUST00000238398.2
|
Lcor
|
ligand dependent nuclear receptor corepressor
|
|
chr10_+_80100868
Show fit
|
1.23 |
ENSMUST00000092305.6
|
Dazap1
|
DAZ associated protein 1
|
|
chr11_+_69804714
Show fit
|
1.22 |
ENSMUST00000072581.9
ENSMUST00000116358.8
|
Gps2
|
G protein pathway suppressor 2
|
|
chr13_+_55359866
Show fit
|
1.21 |
ENSMUST00000224693.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1
|
|
chr3_-_142587419
Show fit
|
1.19 |
ENSMUST00000174422.8
ENSMUST00000173830.8
|
Pkn2
|
protein kinase N2
|
|
chr5_+_115149170
Show fit
|
1.18 |
ENSMUST00000031530.9
|
Sppl3
|
signal peptide peptidase 3
|
|
chr19_+_4806544
Show fit
|
1.17 |
ENSMUST00000182821.8
ENSMUST00000036744.8
|
Rbm4b
|
RNA binding motif protein 4B
|
|
chr14_-_30740946
Show fit
|
1.17 |
ENSMUST00000228341.2
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr11_+_74721733
Show fit
|
1.17 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein
|
|
chr3_-_88204145
Show fit
|
1.16 |
ENSMUST00000010682.4
|
Tsacc
|
TSSK6 activating co-chaperone
|
|
chr2_+_75489596
Show fit
|
1.15 |
ENSMUST00000111964.8
ENSMUST00000111962.8
ENSMUST00000111961.8
ENSMUST00000164947.9
ENSMUST00000090792.11
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chrX_+_85235370
Show fit
|
1.15 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr9_+_56983679
Show fit
|
1.15 |
ENSMUST00000168177.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast)
|
|
chr14_-_52434863
Show fit
|
1.15 |
ENSMUST00000046709.9
|
Supt16
|
SPT16, facilitates chromatin remodeling subunit
|
|
chr19_-_60569323
Show fit
|
1.15 |
ENSMUST00000111460.5
ENSMUST00000166712.9
ENSMUST00000081790.15
|
Cacul1
|
CDK2 associated, cullin domain 1
|
|
chr2_-_37593287
Show fit
|
1.14 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr13_+_55359598
Show fit
|
1.14 |
ENSMUST00000224918.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1
|
|
chr17_-_80203457
Show fit
|
1.12 |
ENSMUST00000068282.7
ENSMUST00000112437.8
|
Atl2
|
atlastin GTPase 2
|
|
chr11_+_114618209
Show fit
|
1.12 |
ENSMUST00000069325.14
|
Dnai2
|
dynein axonemal intermediate chain 2
|
|
chr10_+_69048914
Show fit
|
1.12 |
ENSMUST00000163497.8
ENSMUST00000164212.8
ENSMUST00000067908.14
|
Rhobtb1
|
Rho-related BTB domain containing 1
|
|
chr11_+_106680062
Show fit
|
1.11 |
ENSMUST00000103068.10
ENSMUST00000018516.11
|
Cep95
|
centrosomal protein 95
|
|
chr9_+_56983627
Show fit
|
1.11 |
ENSMUST00000168678.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast)
|
|
chr17_-_36291087
Show fit
|
1.06 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3
|
|
chr12_-_101924407
Show fit
|
1.06 |
ENSMUST00000159883.2
ENSMUST00000160251.8
ENSMUST00000161011.8
ENSMUST00000021606.12
|
Atxn3
|
ataxin 3
|
|
chr14_-_30741012
Show fit
|
1.05 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr11_-_93776580
Show fit
|
1.05 |
ENSMUST00000066888.10
|
Utp18
|
UTP18 small subunit processome component
|
|
chr3_+_107502347
Show fit
|
1.05 |
ENSMUST00000014747.3
|
Alx3
|
aristaless-like homeobox 3
|
|
chr4_-_108690741
Show fit
|
1.04 |
ENSMUST00000102740.8
ENSMUST00000102741.8
|
Btf3l4
|
basic transcription factor 3-like 4
|
|
chr19_+_7034149
Show fit
|
1.04 |
ENSMUST00000040261.7
|
Macrod1
|
mono-ADP ribosylhydrolase 1
|
|
chr19_+_6107874
Show fit
|
1.03 |
ENSMUST00000178310.9
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived)
|
|
chr7_+_89779564
Show fit
|
1.02 |
ENSMUST00000208742.2
ENSMUST00000049537.9
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr9_-_45923908
Show fit
|
1.02 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2
|
|
chr5_-_134485081
Show fit
|
1.01 |
ENSMUST00000111244.5
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1
|
|
chr11_+_57899890
Show fit
|
1.00 |
ENSMUST00000071487.13
ENSMUST00000178636.2
|
Larp1
|
La ribonucleoprotein domain family, member 1
|
|
chr2_+_48839505
Show fit
|
1.00 |
ENSMUST00000112745.8
ENSMUST00000112754.8
|
Mbd5
|
methyl-CpG binding domain protein 5
|
|
chr3_+_41519289
Show fit
|
0.99 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1
|
|
chr7_+_19311212
Show fit
|
0.99 |
ENSMUST00000108453.2
|
Zfp296
|
zinc finger protein 296
|
|
chr4_+_133974246
Show fit
|
0.98 |
ENSMUST00000205501.2
|
Gm30191
|
predicted gene, 30191
|
|
chr3_+_137570248
Show fit
|
0.98 |
ENSMUST00000041045.14
|
H2az1
|
H2A.Z variant histone 1
|
|
chr6_-_70769135
Show fit
|
0.97 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A
|
|
chr16_+_93885775
Show fit
|
0.96 |
ENSMUST00000072182.9
|
Sim2
|
single-minded family bHLH transcription factor 2
|
|
chr15_+_99024394
Show fit
|
0.95 |
ENSMUST00000063517.6
|
Spats2
|
spermatogenesis associated, serine-rich 2
|
|
chr15_-_76540916
Show fit
|
0.94 |
ENSMUST00000229524.2
|
Cyhr1
|
cysteine and histidine rich 1
|
|
chr17_-_36290571
Show fit
|
0.94 |
ENSMUST00000173724.2
ENSMUST00000172900.8
ENSMUST00000174849.8
|
Prr3
|
proline-rich polypeptide 3
|
|
chr6_-_52194414
Show fit
|
0.93 |
ENSMUST00000140316.2
|
Hoxa7
|
homeobox A7
|
|
chr6_-_52135261
Show fit
|
0.93 |
ENSMUST00000000964.6
ENSMUST00000120363.2
|
Hoxa1
|
homeobox A1
|
|
chr17_+_74835290
Show fit
|
0.93 |
ENSMUST00000180037.8
|
Birc6
|
baculoviral IAP repeat-containing 6
|
|
chr5_+_75236250
Show fit
|
0.93 |
ENSMUST00000040477.4
ENSMUST00000160104.3
|
Gsx2
|
GS homeobox 2
|
|
chr2_-_131001916
Show fit
|
0.91 |
ENSMUST00000103188.10
ENSMUST00000133602.8
ENSMUST00000028800.12
|
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene
|
|
chr7_-_26866157
Show fit
|
0.90 |
ENSMUST00000080058.11
|
Egln2
|
egl-9 family hypoxia-inducible factor 2
|
|
chr17_+_35117438
Show fit
|
0.89 |
ENSMUST00000114033.9
ENSMUST00000078061.13
|
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2
|
|
chr3_-_95725944
Show fit
|
0.88 |
ENSMUST00000200164.5
ENSMUST00000090791.8
ENSMUST00000197449.2
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr19_+_18690556
Show fit
|
0.88 |
ENSMUST00000062753.3
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene
|
|
chr2_+_153187729
Show fit
|
0.87 |
ENSMUST00000227428.2
ENSMUST00000109790.2
|
Asxl1
|
ASXL transcriptional regulator 1
|
|
chr17_+_37253802
Show fit
|
0.87 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39
|
|
chr17_+_37253916
Show fit
|
0.86 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39
|
|
chr15_-_96953823
Show fit
|
0.86 |
ENSMUST00000023101.10
|
Slc38a4
|
solute carrier family 38, member 4
|
|
chr10_-_115151451
Show fit
|
0.85 |
ENSMUST00000020343.9
ENSMUST00000218831.2
|
Rab21
|
RAB21, member RAS oncogene family
|
|
chr13_+_9143995
Show fit
|
0.83 |
ENSMUST00000091829.4
|
Larp4b
|
La ribonucleoprotein domain family, member 4B
|
|
chr15_-_76541105
Show fit
|
0.83 |
ENSMUST00000176274.2
|
Cyhr1
|
cysteine and histidine rich 1
|
|
chr13_-_54836077
Show fit
|
0.82 |
ENSMUST00000150626.2
ENSMUST00000134177.8
|
Rnf44
|
ring finger protein 44
|
|
chr17_+_35117905
Show fit
|
0.82 |
ENSMUST00000097342.10
ENSMUST00000013931.12
|
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2
|
|
chr14_+_30741082
Show fit
|
0.81 |
ENSMUST00000112098.11
ENSMUST00000112095.8
ENSMUST00000112106.8
ENSMUST00000146325.8
|
Pbrm1
|
polybromo 1
|
|
chr17_+_46694646
Show fit
|
0.80 |
ENSMUST00000113481.9
ENSMUST00000138127.8
|
Zfp318
|
zinc finger protein 318
|
|
chr2_-_48839276
Show fit
|
0.80 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4
|
|
chr6_-_52194440
Show fit
|
0.80 |
ENSMUST00000048715.9
|
Hoxa7
|
homeobox A7
|
|
chr17_-_48189815
Show fit
|
0.80 |
ENSMUST00000154108.2
|
Foxp4
|
forkhead box P4
|
|
chr19_+_4806859
Show fit
|
0.80 |
ENSMUST00000237921.2
|
Rbm4b
|
RNA binding motif protein 4B
|
|
chr1_-_23961379
Show fit
|
0.77 |
ENSMUST00000027339.14
|
Smap1
|
small ArfGAP 1
|
|
chr17_+_34159633
Show fit
|
0.76 |
ENSMUST00000025170.11
|
Wdr46
|
WD repeat domain 46
|
|
chr2_-_27317004
Show fit
|
0.76 |
ENSMUST00000056176.8
|
Vav2
|
vav 2 oncogene
|
|
chr19_+_18690589
Show fit
|
0.75 |
ENSMUST00000055792.8
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene
|
|
chr11_+_74540284
Show fit
|
0.75 |
ENSMUST00000117818.2
ENSMUST00000092915.12
|
Cluh
|
clustered mitochondria (cluA/CLU1) homolog
|
|
chr4_+_134195631
Show fit
|
0.74 |
ENSMUST00000030636.11
ENSMUST00000127279.8
ENSMUST00000105867.8
|
Stmn1
|
stathmin 1
|
|
chr17_+_36290743
Show fit
|
0.74 |
ENSMUST00000087200.4
|
Gnl1
|
guanine nucleotide binding protein-like 1
|
|
chr3_-_96634880
Show fit
|
0.74 |
ENSMUST00000029741.9
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C
|
|
chr16_+_43960183
Show fit
|
0.73 |
ENSMUST00000159514.8
ENSMUST00000161326.8
ENSMUST00000063520.15
ENSMUST00000063542.8
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr11_-_104333059
Show fit
|
0.73 |
ENSMUST00000106977.8
ENSMUST00000106972.8
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1
|
|
chr2_+_146063841
Show fit
|
0.72 |
ENSMUST00000089257.6
|
Insm1
|
insulinoma-associated 1
|
|
chr11_-_102207516
Show fit
|
0.72 |
ENSMUST00000107115.8
ENSMUST00000128016.2
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I
|
|
chr6_+_119152210
Show fit
|
0.72 |
ENSMUST00000112777.9
ENSMUST00000073909.6
|
Dcp1b
|
decapping mRNA 1B
|
|
chr3_+_88439616
Show fit
|
0.72 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A
|
|
chr11_-_88755360
Show fit
|
0.71 |
ENSMUST00000018572.11
|
Akap1
|
A kinase (PRKA) anchor protein 1
|
|
chr19_-_10181243
Show fit
|
0.71 |
ENSMUST00000142241.2
ENSMUST00000116542.9
ENSMUST00000025651.6
ENSMUST00000156291.2
|
Fen1
|
flap structure specific endonuclease 1
|
|
chr8_+_106075475
Show fit
|
0.71 |
ENSMUST00000073149.7
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5
|
|
chr15_-_79816785
Show fit
|
0.70 |
ENSMUST00000089293.11
ENSMUST00000109616.9
|
Cbx7
|
chromobox 7
|
|
chr7_+_127111576
Show fit
|
0.69 |
ENSMUST00000186672.7
|
Srcap
|
Snf2-related CREBBP activator protein
|
|
chr11_-_51891259
Show fit
|
0.69 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B
|
|
chr3_-_96634752
Show fit
|
0.69 |
ENSMUST00000154679.8
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C
|
|
chr11_-_98040377
Show fit
|
0.69 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20
|
|
chr16_+_29884153
Show fit
|
0.69 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1
|
|
chr8_+_123202882
Show fit
|
0.68 |
ENSMUST00000116412.8
|
Ctu2
|
cytosolic thiouridylase subunit 2
|
|
chr13_-_54835508
Show fit
|
0.67 |
ENSMUST00000177950.8
ENSMUST00000146931.8
|
Rnf44
|
ring finger protein 44
|
|
chr8_+_123202935
Show fit
|
0.67 |
ENSMUST00000146634.8
ENSMUST00000134127.2
|
Ctu2
|
cytosolic thiouridylase subunit 2
|
|
chr11_+_60308077
Show fit
|
0.66 |
ENSMUST00000070681.7
|
Gid4
|
GID complex subunit 4, VID24 homolog
|
|
chr19_-_4447080
Show fit
|
0.65 |
ENSMUST00000075856.11
ENSMUST00000176483.3
ENSMUST00000116571.9
|
Kdm2a
|
lysine (K)-specific demethylase 2A
|
|
chr15_-_79816717
Show fit
|
0.65 |
ENSMUST00000177044.2
ENSMUST00000109615.8
|
Cbx7
|
chromobox 7
|
|
chr9_+_107431776
Show fit
|
0.65 |
ENSMUST00000010211.7
|
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr11_-_102207486
Show fit
|
0.64 |
ENSMUST00000146896.9
ENSMUST00000079589.11
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I
|