Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
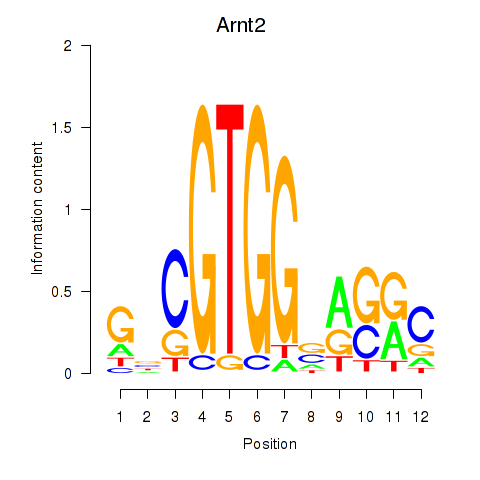
Results for Arnt2
Z-value: 0.35
Transcription factors associated with Arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt2
|
ENSMUSG00000015709.10 | Arnt2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt2 | mm39_v1_chr7_-_84059170_84059210 | 0.75 | 4.4e-14 | Click! |
Activity profile of Arnt2 motif
Sorted Z-values of Arnt2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_101353742 | 4.75 |
ENSMUST00000154120.9
ENSMUST00000106930.8 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr19_-_5148506 | 3.96 |
ENSMUST00000025805.8
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr7_-_45019984 | 3.88 |
ENSMUST00000003971.10
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr19_+_23736205 | 2.83 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr11_+_42310557 | 2.29 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr19_+_22425534 | 1.94 |
ENSMUST00000235522.2
ENSMUST00000236372.2 ENSMUST00000238066.2 ENSMUST00000235780.2 ENSMUST00000236804.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr2_-_131404203 | 1.74 |
ENSMUST00000103184.4
|
Adra1d
|
adrenergic receptor, alpha 1d |
| chr2_-_33261498 | 1.69 |
ENSMUST00000113165.8
|
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr14_-_63781381 | 1.57 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr2_-_33261411 | 1.47 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr9_+_65278979 | 1.39 |
ENSMUST00000239433.2
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr1_-_52856801 | 1.12 |
ENSMUST00000027271.9
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr5_-_135280063 | 1.07 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
| chr1_-_52856662 | 0.99 |
ENSMUST00000162576.8
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr9_-_44231526 | 0.85 |
ENSMUST00000214602.2
ENSMUST00000065080.10 |
C2cd2l
|
C2 calcium-dependent domain containing 2-like |
| chr19_+_22425565 | 0.85 |
ENSMUST00000037901.14
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr5_+_88712840 | 0.83 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr11_-_45835737 | 0.83 |
ENSMUST00000129820.8
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr8_+_84627332 | 0.80 |
ENSMUST00000045393.15
ENSMUST00000132500.8 ENSMUST00000152978.8 |
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr10_+_11219117 | 0.60 |
ENSMUST00000069106.5
|
Epm2a
|
epilepsy, progressive myoclonic epilepsy, type 2 gene alpha |
| chr7_+_18810167 | 0.58 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr4_+_123010035 | 0.57 |
ENSMUST00000102648.6
|
Oxct2b
|
3-oxoacid CoA transferase 2B |
| chr2_+_79085844 | 0.55 |
ENSMUST00000099972.5
|
Itga4
|
integrin alpha 4 |
| chr11_+_107438751 | 0.55 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr7_+_18810097 | 0.44 |
ENSMUST00000032570.14
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr4_-_129534853 | 0.36 |
ENSMUST00000046425.16
ENSMUST00000133803.8 |
Txlna
|
taxilin alpha |
| chr2_+_91480460 | 0.34 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr1_-_59012579 | 0.33 |
ENSMUST00000173590.2
ENSMUST00000027186.12 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr4_-_129534752 | 0.32 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr4_-_123217391 | 0.32 |
ENSMUST00000102640.2
|
Oxct2a
|
3-oxoacid CoA transferase 2A |
| chr7_+_130467486 | 0.21 |
ENSMUST00000120441.8
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr16_-_4238280 | 0.20 |
ENSMUST00000120080.8
|
Adcy9
|
adenylate cyclase 9 |
| chr3_+_96604390 | 0.10 |
ENSMUST00000162778.3
ENSMUST00000064900.16 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr11_+_43419586 | 0.10 |
ENSMUST00000050574.7
|
Ccnjl
|
cyclin J-like |
| chr1_+_74627445 | 0.07 |
ENSMUST00000113733.10
ENSMUST00000027358.11 |
Bcs1l
|
BCS1-like (yeast) |
| chr10_-_121933276 | 0.03 |
ENSMUST00000140299.3
|
Rxylt1
|
ribitol xylosyltransferase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.8 | 4.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.6 | 3.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 1.7 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.3 | 3.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 1.1 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.2 | 0.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 2.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 2.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.6 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.6 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.3 | GO:1903336 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 2.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.8 | GO:0090129 | calcium-mediated signaling using intracellular calcium source(GO:0035584) positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.9 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 0.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 4.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 2.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 5.6 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 2.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.5 | 3.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 1.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 2.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 0.9 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 0.8 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.9 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.0 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |