Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Ascl2
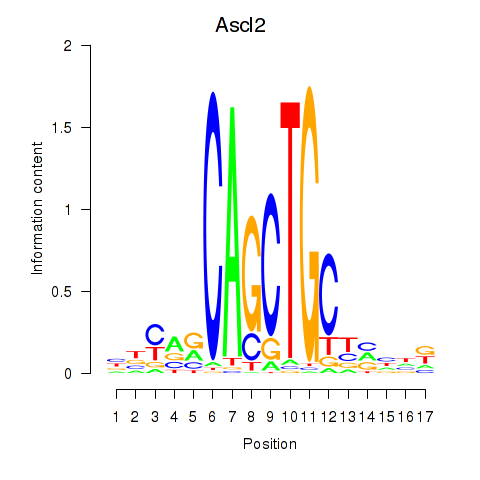
Z-value: 1.75
Transcription factors associated with Ascl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ascl2
|
ENSMUSG00000009248.7 | Ascl2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ascl2 | mm39_v1_chr7_-_142522995_142523006 | 0.23 | 5.4e-02 | Click! |
Activity profile of Ascl2 motif
Sorted Z-values of Ascl2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ascl2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_111608339 | 17.33 |
ENSMUST00000039887.4
|
Pof1b
|
premature ovarian failure 1B |
| chr5_-_98178811 | 16.81 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2 |
| chr16_+_57173456 | 15.21 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_+_142025575 | 12.70 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr12_-_103423472 | 12.37 |
ENSMUST00000044687.7
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr13_-_63006176 | 12.00 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr12_-_114752425 | 11.97 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr5_+_136996713 | 11.36 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr3_-_100396635 | 10.29 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr7_+_142025817 | 10.12 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr11_+_62711057 | 9.92 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr4_-_43523745 | 9.90 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr4_-_43523595 | 9.32 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr11_+_69856222 | 9.20 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr5_-_134975773 | 9.13 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr15_+_101310283 | 9.11 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr10_-_128237087 | 8.62 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr2_+_153334710 | 8.35 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr6_-_69741999 | 8.27 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr9_-_71499628 | 8.09 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr15_-_74983786 | 7.75 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr7_+_43874854 | 7.63 |
ENSMUST00000206144.2
|
Klk1
|
kallikrein 1 |
| chr2_-_179976458 | 7.50 |
ENSMUST00000015771.3
|
Gata5
|
GATA binding protein 5 |
| chr2_-_147888816 | 7.48 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr9_-_71499547 | 7.47 |
ENSMUST00000169573.8
|
Myzap
|
myocardial zonula adherens protein |
| chr4_-_141345549 | 7.42 |
ENSMUST00000053263.9
|
Tmem82
|
transmembrane protein 82 |
| chr7_+_27770655 | 7.38 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr12_-_114878652 | 7.36 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr7_-_127593003 | 7.34 |
ENSMUST00000033056.5
|
Pycard
|
PYD and CARD domain containing |
| chr1_+_177270101 | 7.18 |
ENSMUST00000194319.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr19_+_6155804 | 7.16 |
ENSMUST00000044451.4
|
Naaladl1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr11_+_99755302 | 7.11 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr4_-_133600308 | 6.97 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr4_-_43523388 | 6.95 |
ENSMUST00000107913.10
ENSMUST00000030184.12 |
Tpm2
|
tropomyosin 2, beta |
| chr6_-_69835868 | 6.88 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr11_+_62711295 | 6.75 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr19_+_38384428 | 6.75 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr12_-_103409912 | 6.70 |
ENSMUST00000055071.9
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr1_+_171246593 | 6.64 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr9_-_45115381 | 6.63 |
ENSMUST00000034599.15
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr15_+_101936615 | 6.58 |
ENSMUST00000023803.8
|
Krt18
|
keratin 18 |
| chr8_-_112417633 | 6.51 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr1_+_172327812 | 6.50 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr2_+_122479770 | 6.48 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr7_-_44180700 | 6.43 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chrX_+_100492684 | 6.16 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr5_+_91287448 | 6.16 |
ENSMUST00000031325.6
|
Areg
|
amphiregulin |
| chr11_+_96820220 | 6.14 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr11_+_117700479 | 6.10 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr6_-_68681962 | 6.06 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr6_+_135339543 | 5.94 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr5_-_98178834 | 5.93 |
ENSMUST00000199088.2
|
Antxr2
|
anthrax toxin receptor 2 |
| chr16_+_57173632 | 5.88 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_+_3339059 | 5.79 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr6_+_135339929 | 5.72 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_83007635 | 5.71 |
ENSMUST00000037994.8
|
Slfn1
|
schlafen 1 |
| chr7_+_3339077 | 5.64 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_-_80037153 | 5.60 |
ENSMUST00000206728.2
|
Fes
|
feline sarcoma oncogene |
| chr1_-_133352115 | 5.52 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr16_+_65612083 | 5.52 |
ENSMUST00000168064.3
|
Vgll3
|
vestigial like family member 3 |
| chr11_+_121593219 | 5.49 |
ENSMUST00000036742.14
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr12_+_113104085 | 5.47 |
ENSMUST00000200380.5
|
Crip2
|
cysteine rich protein 2 |
| chr16_+_65612394 | 5.47 |
ENSMUST00000227997.2
|
Vgll3
|
vestigial like family member 3 |
| chr11_+_117740077 | 5.43 |
ENSMUST00000081387.11
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr14_+_34395845 | 5.42 |
ENSMUST00000048263.14
|
Wapl
|
WAPL cohesin release factor |
| chr1_-_135846937 | 5.39 |
ENSMUST00000027667.13
|
Pkp1
|
plakophilin 1 |
| chr1_-_156501860 | 5.35 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr5_+_35198853 | 5.35 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr3_+_106393348 | 5.34 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr12_-_113733922 | 5.30 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr6_-_52168675 | 5.27 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr12_-_115916604 | 5.19 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr17_-_35285146 | 5.13 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr12_-_115832846 | 5.06 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr12_-_113790741 | 5.06 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr12_+_113103817 | 5.02 |
ENSMUST00000084882.9
|
Crip2
|
cysteine rich protein 2 |
| chr1_+_40720731 | 4.96 |
ENSMUST00000192345.2
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr17_-_57535003 | 4.96 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr11_-_99519057 | 4.95 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr14_-_70761507 | 4.95 |
ENSMUST00000022692.5
|
Sftpc
|
surfactant associated protein C |
| chr15_+_75027089 | 4.94 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr2_-_174280811 | 4.91 |
ENSMUST00000016400.9
|
Ctsz
|
cathepsin Z |
| chr1_-_167221344 | 4.83 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr7_-_141023199 | 4.82 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr9_+_107784065 | 4.77 |
ENSMUST00000035203.9
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr7_-_28246530 | 4.75 |
ENSMUST00000239002.2
ENSMUST00000057974.4 |
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr1_-_52539395 | 4.68 |
ENSMUST00000186764.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr4_-_46536088 | 4.67 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr1_-_135846858 | 4.66 |
ENSMUST00000163260.8
|
Pkp1
|
plakophilin 1 |
| chr17_-_43813664 | 4.63 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr8_+_96078886 | 4.58 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr6_+_127538268 | 4.52 |
ENSMUST00000212051.2
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr9_-_66951151 | 4.49 |
ENSMUST00000113696.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr7_+_43874752 | 4.44 |
ENSMUST00000075162.5
|
Klk1
|
kallikrein 1 |
| chr9_-_71499594 | 4.41 |
ENSMUST00000166112.2
|
Myzap
|
myocardial zonula adherens protein |
| chr16_+_78727829 | 4.38 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr9_-_66951025 | 4.37 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr16_-_36648586 | 4.36 |
ENSMUST00000023537.6
ENSMUST00000114829.9 |
Eaf2
|
ELL associated factor 2 |
| chr13_+_54849268 | 4.34 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr2_-_25086810 | 4.33 |
ENSMUST00000081869.7
|
Tor4a
|
torsin family 4, member A |
| chr17_+_87590308 | 4.32 |
ENSMUST00000041110.12
ENSMUST00000125875.8 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr17_+_48096628 | 4.29 |
ENSMUST00000024786.14
|
Tfeb
|
transcription factor EB |
| chr2_+_167380112 | 4.28 |
ENSMUST00000052631.8
|
Snai1
|
snail family zinc finger 1 |
| chr11_+_33913013 | 4.27 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr11_+_98337655 | 4.26 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr7_+_89053562 | 4.25 |
ENSMUST00000058755.5
|
Fzd4
|
frizzled class receptor 4 |
| chr11_-_106240215 | 4.22 |
ENSMUST00000021056.8
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha |
| chr5_+_64317550 | 4.21 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr9_+_110867807 | 4.20 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr5_-_107873883 | 4.18 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr4_-_131695135 | 4.18 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr2_+_164245114 | 4.17 |
ENSMUST00000017151.2
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr18_+_64473091 | 4.16 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr9_+_44893077 | 4.15 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr14_-_56181993 | 4.14 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr19_-_41373526 | 4.08 |
ENSMUST00000059672.9
|
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr6_+_70680515 | 4.05 |
ENSMUST00000103404.2
|
Igkv3-1
|
immunoglobulin kappa variable 3-1 |
| chr18_-_32692967 | 4.01 |
ENSMUST00000174000.2
ENSMUST00000174459.2 |
Gypc
|
glycophorin C |
| chr7_-_80037622 | 3.94 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr4_-_63965161 | 3.90 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr7_+_44866095 | 3.88 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr12_+_79177523 | 3.87 |
ENSMUST00000021550.7
|
Arg2
|
arginase type II |
| chr11_+_99805555 | 3.87 |
ENSMUST00000105053.2
|
Gm11565
|
predicted gene 11565 |
| chr5_-_100867520 | 3.87 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr15_-_89263448 | 3.85 |
ENSMUST00000049968.9
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr12_+_71063431 | 3.84 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr8_-_41827639 | 3.83 |
ENSMUST00000034000.15
ENSMUST00000143057.2 |
Asah1
|
N-acylsphingosine amidohydrolase 1 |
| chr5_-_66330394 | 3.82 |
ENSMUST00000201544.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr7_-_45173193 | 3.80 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr7_+_75105282 | 3.79 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr2_-_25086770 | 3.79 |
ENSMUST00000142857.2
ENSMUST00000137920.2 |
Tor4a
|
torsin family 4, member A |
| chr11_+_83067429 | 3.76 |
ENSMUST00000215472.2
|
Slfn4
|
schlafen 4 |
| chr2_+_35172392 | 3.76 |
ENSMUST00000028239.8
|
Gsn
|
gelsolin |
| chr5_-_28415020 | 3.75 |
ENSMUST00000118882.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr2_-_60383647 | 3.75 |
ENSMUST00000112525.5
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr18_-_10182007 | 3.71 |
ENSMUST00000067947.7
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr1_+_174000304 | 3.71 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr7_+_142014546 | 3.66 |
ENSMUST00000105968.8
ENSMUST00000018963.11 ENSMUST00000105967.8 |
Lsp1
|
lymphocyte specific 1 |
| chr8_-_71834543 | 3.66 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr4_-_123236596 | 3.63 |
ENSMUST00000102641.10
|
Bmp8a
|
bone morphogenetic protein 8a |
| chr15_-_89263790 | 3.60 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr5_+_123153072 | 3.58 |
ENSMUST00000051016.5
ENSMUST00000121652.8 |
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr6_+_68495964 | 3.58 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr11_+_117740111 | 3.58 |
ENSMUST00000093906.5
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr3_+_108272205 | 3.54 |
ENSMUST00000090563.7
|
Mybphl
|
myosin binding protein H-like |
| chr10_+_75729237 | 3.53 |
ENSMUST00000009236.6
ENSMUST00000217811.2 |
Derl3
|
Der1-like domain family, member 3 |
| chr19_+_7034149 | 3.50 |
ENSMUST00000040261.7
|
Macrod1
|
mono-ADP ribosylhydrolase 1 |
| chr19_+_37423198 | 3.50 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr6_+_112436466 | 3.50 |
ENSMUST00000075477.8
|
Cav3
|
caveolin 3 |
| chr10_+_34359395 | 3.48 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr9_+_107852733 | 3.46 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr17_-_45906768 | 3.45 |
ENSMUST00000164618.8
ENSMUST00000097317.10 ENSMUST00000170113.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr11_+_98798627 | 3.43 |
ENSMUST00000092706.13
|
Cdc6
|
cell division cycle 6 |
| chr4_-_147894245 | 3.43 |
ENSMUST00000105734.10
ENSMUST00000176201.2 |
Zfp984
Gm20707
|
zinc finger protein 984 predicted gene 20707 |
| chr7_+_44866635 | 3.43 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr8_-_106198112 | 3.42 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr4_-_132990362 | 3.35 |
ENSMUST00000105908.10
ENSMUST00000030674.8 |
Sytl1
|
synaptotagmin-like 1 |
| chr15_-_89263128 | 3.35 |
ENSMUST00000227834.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr7_-_103964662 | 3.34 |
ENSMUST00000106837.8
ENSMUST00000106839.9 ENSMUST00000070943.7 |
Trim12a
|
tripartite motif-containing 12A |
| chr16_+_48637219 | 3.33 |
ENSMUST00000023328.8
|
Retnlb
|
resistin like beta |
| chr7_-_127805518 | 3.33 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr5_-_106606032 | 3.32 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr15_+_102008829 | 3.32 |
ENSMUST00000228958.2
|
Tns2
|
tensin 2 |
| chr4_-_133599616 | 3.31 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_+_45871135 | 3.30 |
ENSMUST00000049038.4
|
Sox30
|
SRY (sex determining region Y)-box 30 |
| chr7_+_107264518 | 3.28 |
ENSMUST00000239294.2
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr10_+_34359513 | 3.28 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr12_-_113386312 | 3.24 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chr7_-_79765042 | 3.23 |
ENSMUST00000206714.2
ENSMUST00000107384.10 |
Idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chrX_-_166047275 | 3.23 |
ENSMUST00000112170.2
|
Tlr8
|
toll-like receptor 8 |
| chr11_+_61099447 | 3.23 |
ENSMUST00000108716.8
ENSMUST00000019246.4 |
Aldh3a1
|
aldehyde dehydrogenase family 3, subfamily A1 |
| chr10_-_128796834 | 3.20 |
ENSMUST00000026398.5
|
Mettl7b
|
methyltransferase like 7B |
| chr9_-_114673158 | 3.19 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr15_-_101588714 | 3.19 |
ENSMUST00000023786.7
|
Krt6b
|
keratin 6B |
| chr4_-_137157824 | 3.19 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr13_-_92024994 | 3.18 |
ENSMUST00000022122.4
|
Ckmt2
|
creatine kinase, mitochondrial 2 |
| chr19_-_45548942 | 3.18 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr15_-_75760602 | 3.17 |
ENSMUST00000184858.2
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr7_+_78563964 | 3.17 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr11_+_98555167 | 3.15 |
ENSMUST00000017348.3
|
Gsdma
|
gasdermin A |
| chr2_+_127696548 | 3.14 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr19_-_41836514 | 3.12 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr7_-_103937301 | 3.12 |
ENSMUST00000098179.9
|
Trim5
|
tripartite motif-containing 5 |
| chr15_-_74855264 | 3.11 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr2_-_92201342 | 3.11 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr18_+_32055339 | 3.11 |
ENSMUST00000233994.2
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr6_+_125326662 | 3.10 |
ENSMUST00000032491.15
|
Tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr19_-_6014210 | 3.10 |
ENSMUST00000025752.15
ENSMUST00000165143.3 |
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr5_+_137639538 | 3.09 |
ENSMUST00000177466.8
ENSMUST00000166099.3 |
Sap25
|
sin3 associated polypeptide |
| chr15_-_89263466 | 3.09 |
ENSMUST00000228111.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr5_+_87148697 | 3.08 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr9_-_66950991 | 3.05 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr2_+_33106062 | 3.04 |
ENSMUST00000004208.7
|
Angptl2
|
angiopoietin-like 2 |
| chr12_-_113680723 | 3.02 |
ENSMUST00000179657.2
|
Ighv2-6
|
immunoglobulin heavy variable 2-6 |
| chr1_+_87254729 | 3.00 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr16_+_93629009 | 2.99 |
ENSMUST00000044068.10
ENSMUST00000202261.5 ENSMUST00000201097.3 |
Morc3
|
microrchidia 3 |
| chr19_-_57185988 | 2.98 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr12_-_115410489 | 2.96 |
ENSMUST00000194581.2
|
Ighv1-62-2
|
immunoglobulin heavy variable 1-62-2 |
| chr7_+_78564062 | 2.95 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr11_-_115426618 | 2.93 |
ENSMUST00000121185.8
ENSMUST00000117589.8 |
Sumo2
|
small ubiquitin-like modifier 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 2.6 | 15.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.5 | 7.5 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 2.4 | 7.3 | GO:0002582 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 1.9 | 5.7 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 1.7 | 8.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.6 | 10.9 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) |
| 1.5 | 4.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 1.5 | 6.2 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.4 | 10.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.4 | 11.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.4 | 4.3 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 1.4 | 17.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.4 | 5.4 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.3 | 3.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 1.3 | 9.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 1.3 | 3.8 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 1.2 | 5.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.2 | 3.5 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 1.1 | 10.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 1.1 | 3.2 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.1 | 3.2 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 1.1 | 4.3 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 1.1 | 7.5 | GO:0015862 | uridine transport(GO:0015862) |
| 1.1 | 4.2 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.0 | 4.1 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 1.0 | 6.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.0 | 3.9 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) bud outgrowth involved in lung branching(GO:0060447) |
| 1.0 | 6.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.0 | 2.9 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.9 | 10.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.9 | 3.8 | GO:0097017 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.9 | 2.8 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.9 | 4.3 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.8 | 2.5 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.8 | 2.5 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.8 | 3.9 | GO:0090467 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.7 | 3.7 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.7 | 2.9 | GO:2000832 | testosterone secretion(GO:0035936) negative regulation of steroid hormone secretion(GO:2000832) regulation of testosterone secretion(GO:2000843) |
| 0.7 | 2.2 | GO:0002851 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.7 | 3.5 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.7 | 2.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.7 | 4.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.7 | 2.7 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.7 | 2.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.6 | 3.9 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.6 | 1.9 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.6 | 6.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.6 | 3.7 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.6 | 2.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.6 | 1.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.6 | 1.8 | GO:0070318 | myoblast development(GO:0048627) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.6 | 1.8 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.6 | 3.5 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.6 | 1.8 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.6 | 4.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.6 | 2.3 | GO:0072190 | ureter urothelium development(GO:0072190) |
| 0.6 | 4.5 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.6 | 2.3 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.6 | 3.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.5 | 1.6 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.5 | 4.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 3.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 3.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.5 | 4.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.5 | 3.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.5 | 3.5 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 0.5 | 4.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.5 | 3.8 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.5 | 5.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 9.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.5 | 1.4 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.5 | 5.0 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 1.8 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.4 | 10.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.4 | 1.3 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.4 | 3.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.4 | 1.3 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.4 | 3.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 9.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.4 | 0.8 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.4 | 2.8 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 4.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.4 | 3.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 11.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 2.6 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 22.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.4 | 2.6 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.4 | 4.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.4 | 9.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.0 | GO:0061723 | glycophagy(GO:0061723) |
| 0.3 | 2.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 1.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 1.7 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 4.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 7.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.3 | 5.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 2.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 2.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 50.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.3 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 3.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 3.7 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.3 | 5.6 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.3 | 4.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.3 | 1.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 3.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 1.2 | GO:0072186 | lactic acid secretion(GO:0046722) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 | 0.9 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.3 | 19.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.3 | 3.8 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.3 | 1.6 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.3 | 2.6 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.3 | 6.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 1.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 1.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 | 6.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 1.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 1.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.2 | 1.0 | GO:0046340 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 2.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 4.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 7.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 1.4 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 0.9 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 4.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 3.6 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.2 | 11.3 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.2 | 9.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 3.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 3.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.2 | 0.8 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 2.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 0.6 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 2.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 2.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 4.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 5.0 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 0.4 | GO:0051464 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of cortisol secretion(GO:0051464) |
| 0.2 | 3.7 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 1.8 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.2 | 2.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 0.5 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.2 | 3.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 1.5 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.2 | 2.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 2.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 1.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 1.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 6.4 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 3.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 1.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 3.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 0.6 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 4.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.6 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.9 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 2.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 1.0 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 0.6 | GO:0090345 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 1.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 1.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 2.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 6.0 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 2.8 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 1.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 2.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 3.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 2.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 1.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.4 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 1.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 3.0 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 1.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 4.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.7 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.1 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) cellular response to morphine(GO:0071315) |
| 0.1 | 3.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 1.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.7 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.1 | 16.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 2.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 4.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.1 | GO:0003195 | tricuspid valve formation(GO:0003195) |
| 0.1 | 0.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 5.3 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 2.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 2.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 2.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.8 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 2.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.4 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.7 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 10.2 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 2.6 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 2.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.9 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.2 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 3.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 1.9 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 1.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 6.4 | GO:0051302 | regulation of cell division(GO:0051302) |
| 0.0 | 1.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 2.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 1.3 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 3.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.9 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 1.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.9 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0040016 | negative regulation of superoxide anion generation(GO:0032929) embryonic cleavage(GO:0040016) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.7 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.9 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 2.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.4 | GO:0007015 | actin filament organization(GO:0007015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 41.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 2.3 | 9.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.8 | 7.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 1.8 | 7.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.5 | 4.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.8 | 5.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.8 | 26.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.7 | 8.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.7 | 4.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.5 | 3.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 2.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.5 | 9.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 3.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.5 | 1.9 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.5 | 15.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.5 | 5.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 53.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 4.3 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 4.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 16.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 3.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 3.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.3 | 2.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 34.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 3.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 3.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 3.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 17.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.2 | 1.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.2 | 3.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 1.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 1.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 4.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 2.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 3.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 3.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 19.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 11.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 8.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 4.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 2.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 6.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 11.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 2.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 19.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 3.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 7.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 2.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 35.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 6.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 4.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 21.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 2.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 23.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 6.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 1.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 9.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 6.9 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.8 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 2.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 27.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 1.9 | 7.5 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.5 | 4.6 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 1.5 | 5.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.3 | 16.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 1.2 | 6.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.0 | 3.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.8 | 2.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 3.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.8 | 7.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 12.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.8 | 3.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.8 | 2.3 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.7 | 5.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.7 | 2.9 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.7 | 2.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.7 | 3.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.7 | 46.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.6 | 2.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.6 | 3.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.6 | 1.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.6 | 2.9 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.6 | 1.7 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.6 | 3.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.6 | 2.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.5 | 2.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 5.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 2.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.5 | 1.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.5 | 1.9 | GO:0016822 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.5 | 63.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.5 | 1.4 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.5 | 3.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 1.4 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.4 | 1.3 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.4 | 1.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 4.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 1.7 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 1.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 1.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.4 | 4.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 1.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 3.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.3 | 2.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 10.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 3.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 2.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 1.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 9.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 2.3 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.3 | 3.7 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 2.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 3.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 1.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 1.2 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.3 | 7.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 3.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 18.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 5.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 3.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 8.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 1.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 6.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 3.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 0.8 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.2 | 1.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 9.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 3.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 0.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.6 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 0.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 7.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 12.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 3.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 2.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 3.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 38.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 47.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 0.6 | GO:0051538 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 2.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 3.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.7 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 0.9 | GO:0043546 | molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 4.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 4.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 3.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 6.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 3.9 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 1.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 7.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 3.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 4.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.6 | GO:0016675 | oxidoreductase activity, acting on a heme group of donors(GO:0016675) |
| 0.1 | 0.9 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.6 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 3.1 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 4.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 4.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 2.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 4.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 2.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 8.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 6.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 2.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 5.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 2.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 4.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 2.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 9.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 6.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 5.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.0 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 6.2 | GO:0001067 | regulatory region DNA binding(GO:0000975) regulatory region nucleic acid binding(GO:0001067) transcription regulatory region DNA binding(GO:0044212) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.3 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 1.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 4.3 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.1 | 3.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 2.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 2.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 3.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 3.7 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 29.5 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 6.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 2.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 4.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.5 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 22.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 1.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 2.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 7.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 17.5 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.1 | GO:0004985 | melanocortin receptor activity(GO:0004977) opioid receptor activity(GO:0004985) |
| 0.0 | 1.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 2.9 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 6.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 22.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.6 | 24.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 3.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 8.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 13.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 5.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 6.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 4.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 3.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 9.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 6.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 6.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 4.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 15.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 4.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 3.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 12.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 10.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 4.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 6.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 0.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 13.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 4.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 7.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 39.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 18.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 23.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.5 | 12.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 7.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 7.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 7.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 8.9 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.4 | 4.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 1.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 5.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 3.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 5.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 8.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 8.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 3.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 13.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 6.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 12.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.2 | 10.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 2.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 2.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 10.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 8.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 3.5 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 5.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 3.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 3.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 4.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 4.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 3.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 7.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 7.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 4.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 9.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 5.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.9 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 9.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 6.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 4.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 5.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 4.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 3.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |