Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
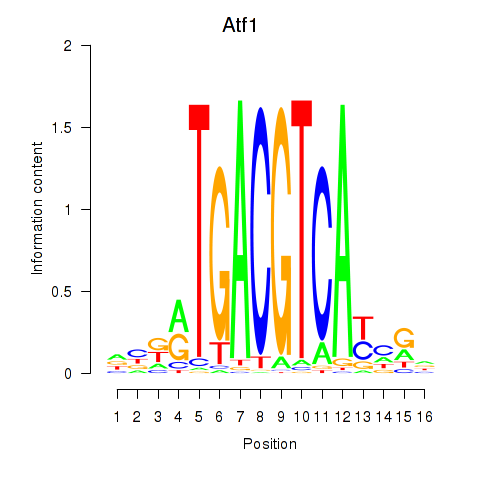
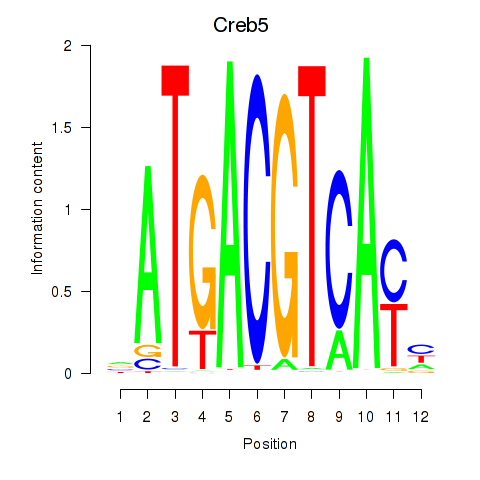
Results for Atf1_Creb5
Z-value: 1.52
Transcription factors associated with Atf1_Creb5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf1
|
ENSMUSG00000023027.14 | Atf1 |
|
Creb5
|
ENSMUSG00000053007.10 | Creb5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb5 | mm39_v1_chr6_+_53550285_53550334 | -0.59 | 5.0e-08 | Click! |
| Atf1 | mm39_v1_chr15_+_100126107_100126163 | 0.44 | 1.3e-04 | Click! |
Activity profile of Atf1_Creb5 motif
Sorted Z-values of Atf1_Creb5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf1_Creb5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_128154709 | 22.57 |
ENSMUST00000053830.5
|
Hmgb4
|
high-mobility group box 4 |
| chr9_-_106438798 | 19.15 |
ENSMUST00010126732.2
ENSMUST00010126033.2 ENSMUST00010181659.1 ENSMUST00010126065.2 ENSMUST00010126032.3 ENSMUST00000062917.16 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr17_+_33651864 | 15.90 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr4_+_43983472 | 14.50 |
ENSMUST00000095107.3
|
Ccin
|
calicin |
| chr17_-_56819422 | 14.36 |
ENSMUST00000052211.4
|
Znrf4
|
zinc and ring finger 4 |
| chr14_+_52155874 | 14.32 |
ENSMUST00000008957.13
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr4_+_56743407 | 13.46 |
ENSMUST00000095079.6
|
Actl7a
|
actin-like 7a |
| chr2_+_18703797 | 13.34 |
ENSMUST00000095132.10
|
Spag6
|
sperm associated antigen 6 |
| chr13_-_23945189 | 13.26 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr4_-_58009118 | 13.02 |
ENSMUST00000102897.11
ENSMUST00000239406.2 |
Txndc8
|
thioredoxin domain containing 8 |
| chr14_-_101437750 | 12.95 |
ENSMUST00000187304.2
|
Prr30
|
proline rich 30 |
| chr7_-_103778992 | 12.53 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr8_-_13612397 | 12.27 |
ENSMUST00000187391.7
ENSMUST00000134023.9 ENSMUST00000151400.10 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr7_+_45271229 | 11.91 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr4_+_102843540 | 11.84 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr5_-_103803578 | 11.59 |
ENSMUST00000120688.6
ENSMUST00000134926.8 ENSMUST00000120108.8 |
1700016H13Rik
|
RIKEN cDNA 1700016H13 gene |
| chr5_+_93354377 | 11.45 |
ENSMUST00000031330.5
|
2010109A12Rik
|
RIKEN cDNA 2010109A12 gene |
| chr2_+_18703863 | 11.38 |
ENSMUST00000173763.2
|
Spag6
|
sperm associated antigen 6 |
| chr16_-_16647139 | 11.34 |
ENSMUST00000023468.6
|
Spag6l
|
sperm associated antigen 6-like |
| chr4_+_51216645 | 11.04 |
ENSMUST00000166749.2
ENSMUST00000156384.4 |
Cylc2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr7_-_24134919 | 10.99 |
ENSMUST00000080594.8
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr19_+_5385672 | 10.97 |
ENSMUST00000043380.5
|
Catsper1
|
cation channel, sperm associated 1 |
| chr2_-_18001734 | 10.96 |
ENSMUST00000105001.4
|
H2al2a
|
H2A histone family member L2A |
| chr12_-_64521464 | 10.89 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr11_-_51153767 | 10.78 |
ENSMUST00000065950.10
|
BC049762
|
cDNA sequence BC049762 |
| chr3_+_134918298 | 10.75 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr9_+_106391771 | 10.69 |
ENSMUST00000085113.5
|
Iqcf5
|
IQ motif containing F5 |
| chr7_+_44145987 | 10.34 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr7_-_126430636 | 10.28 |
ENSMUST00000205320.2
ENSMUST00000061695.5 |
4930451I11Rik
|
RIKEN cDNA 4930451I11 gene |
| chr12_-_28632514 | 10.20 |
ENSMUST00000110917.2
ENSMUST00000020965.14 |
Allc
|
allantoicase |
| chr5_+_25386452 | 10.13 |
ENSMUST00000030778.9
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr13_-_55563028 | 9.94 |
ENSMUST00000054146.5
|
Pfn3
|
profilin 3 |
| chr9_+_106377153 | 9.89 |
ENSMUST00000164965.3
|
Iqcf1
|
IQ motif containing F1 |
| chr9_+_106377181 | 9.75 |
ENSMUST00000085114.8
|
Iqcf1
|
IQ motif containing F1 |
| chr17_+_48400153 | 9.63 |
ENSMUST00000233043.2
|
1700067P10Rik
|
RIKEN cDNA 1700067P10 gene |
| chr1_-_56676589 | 9.36 |
ENSMUST00000062085.6
|
Hsfy2
|
heat shock transcription factor, Y-linked 2 |
| chr7_+_44146029 | 9.24 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr5_+_25386487 | 9.07 |
ENSMUST00000114965.2
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr2_+_172314433 | 8.93 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr18_-_3280999 | 8.85 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr7_+_44926925 | 8.72 |
ENSMUST00000210861.2
|
Slc6a21
|
solute carrier family 6 member 21 |
| chr7_+_44146012 | 8.66 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chrX_-_117390434 | 8.65 |
ENSMUST00000073857.6
|
Tgif2lx1
|
TGFB-induced factor homeobox 2-like, X-linked 1 |
| chr12_-_81426238 | 8.40 |
ENSMUST00000062182.8
|
Gm4787
|
predicted gene 4787 |
| chr1_+_133291302 | 8.30 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr1_+_86354045 | 8.13 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr11_+_87457544 | 8.02 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr6_-_135231324 | 8.00 |
ENSMUST00000111911.9
ENSMUST00000111910.4 |
Gsg1
|
germ cell associated 1 |
| chrX_+_14077387 | 7.95 |
ENSMUST00000105137.4
|
H2al1n
|
H2A histone family member L1N |
| chr15_-_99185050 | 7.78 |
ENSMUST00000109100.2
|
Fam186b
|
family with sequence similarity 186, member B |
| chr6_+_41661356 | 7.55 |
ENSMUST00000031900.6
|
Llcfc1
|
LLLL and CFNLAS motif containing 1 |
| chr4_-_56741398 | 7.53 |
ENSMUST00000095080.5
|
Actl7b
|
actin-like 7b |
| chr4_-_25242858 | 7.31 |
ENSMUST00000029922.14
ENSMUST00000108204.2 |
Fhl5
|
four and a half LIM domains 5 |
| chr2_-_35257741 | 7.18 |
ENSMUST00000028243.2
|
4930568D16Rik
|
RIKEN cDNA 4930568D16 gene |
| chr3_-_107462378 | 7.13 |
ENSMUST00000052853.8
|
Ubl4b
|
ubiquitin-like 4B |
| chr12_-_110649040 | 7.08 |
ENSMUST00000222915.2
ENSMUST00000070659.7 |
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr8_-_105019806 | 6.93 |
ENSMUST00000212492.2
ENSMUST00000034344.10 |
Cmtm2a
|
CKLF-like MARVEL transmembrane domain containing 2A |
| chr8_+_117648474 | 6.85 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr6_+_126830102 | 6.74 |
ENSMUST00000202878.4
ENSMUST00000202574.2 |
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr8_-_26087552 | 6.73 |
ENSMUST00000210234.2
ENSMUST00000211422.2 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr18_-_3281089 | 6.72 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr3_+_55047453 | 6.70 |
ENSMUST00000118963.9
ENSMUST00000061099.14 ENSMUST00000153009.8 |
Ccdc169
|
coiled-coil domain containing 169 |
| chr6_+_88199250 | 6.63 |
ENSMUST00000061866.6
|
Dnajb8
|
DnaJ heat shock protein family (Hsp40) member B8 |
| chr8_-_26087475 | 6.60 |
ENSMUST00000210810.2
ENSMUST00000210616.2 ENSMUST00000079160.8 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr6_-_135231168 | 6.55 |
ENSMUST00000111909.8
|
Gsg1
|
germ cell associated 1 |
| chr9_-_106448182 | 6.50 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr10_-_30076543 | 6.49 |
ENSMUST00000099985.6
|
Cenpw
|
centromere protein W |
| chr5_+_25721059 | 6.40 |
ENSMUST00000045016.9
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chrX_+_9150003 | 6.39 |
ENSMUST00000073949.4
|
H2al1m
|
H2A histone family member L1M |
| chr3_-_127346882 | 6.31 |
ENSMUST00000197668.2
ENSMUST00000029588.10 |
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr14_+_43116106 | 6.19 |
ENSMUST00000169587.2
|
Gm8126
|
predicted gene 8126 |
| chr5_-_143211632 | 6.17 |
ENSMUST00000085733.9
|
Spdye4a
|
speedy/RINGO cell cycle regulator family, member E4A |
| chr2_-_58990967 | 6.17 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chr3_-_145813802 | 6.15 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3 |
| chr2_-_120439764 | 6.12 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr6_+_126830050 | 6.06 |
ENSMUST00000095440.9
|
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr1_-_183766195 | 5.93 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr11_+_87295860 | 5.91 |
ENSMUST00000060835.12
|
Tex14
|
testis expressed gene 14 |
| chr18_+_35987733 | 5.83 |
ENSMUST00000235337.2
|
Cxxc5
|
CXXC finger 5 |
| chr2_-_164587836 | 5.81 |
ENSMUST00000109328.8
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr17_-_29768531 | 5.80 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chrX_+_9715942 | 5.77 |
ENSMUST00000057113.3
|
H2al3
|
H2A histone family member L3 |
| chr17_-_36291087 | 5.75 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3 |
| chr4_-_133972890 | 5.59 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr9_-_106421834 | 5.59 |
ENSMUST00010181660.1
ENSMUST00000215525.2 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr8_+_26339646 | 5.55 |
ENSMUST00000098858.11
|
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr17_+_37269513 | 5.44 |
ENSMUST00000173814.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr3_-_124374723 | 5.43 |
ENSMUST00000180162.8
ENSMUST00000047110.14 ENSMUST00000178485.8 |
1700003H04Rik
|
RIKEN cDNA 1700003H04 gene |
| chr7_-_30371021 | 5.39 |
ENSMUST00000051495.7
|
Pmis2
|
PMIS2 transmembrane protein |
| chr8_+_11890474 | 5.39 |
ENSMUST00000033909.14
ENSMUST00000209692.2 |
Tex29
|
testis expressed 29 |
| chr15_+_73706410 | 5.38 |
ENSMUST00000154520.2
|
Gm6569
|
predicted gene 6569 |
| chr7_-_101859379 | 5.30 |
ENSMUST00000210682.2
|
Nup98
|
nucleoporin 98 |
| chr7_-_30443106 | 5.28 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr7_+_138448061 | 5.24 |
ENSMUST00000041097.13
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr12_+_98594388 | 5.21 |
ENSMUST00000048402.12
ENSMUST00000101144.10 ENSMUST00000101146.4 |
Spata7
|
spermatogenesis associated 7 |
| chr3_+_40754489 | 5.18 |
ENSMUST00000203295.3
|
Plk4
|
polo like kinase 4 |
| chr17_-_56607286 | 5.14 |
ENSMUST00000097303.3
|
Arrdc5
|
arrestin domain containing 5 |
| chr6_+_41098273 | 5.09 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr7_+_138448308 | 5.07 |
ENSMUST00000155672.8
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr6_+_129510331 | 5.02 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr5_-_124170305 | 5.00 |
ENSMUST00000040967.9
|
Vps37b
|
vacuolar protein sorting 37B |
| chr18_+_50411431 | 4.91 |
ENSMUST00000039121.4
ENSMUST00000238078.2 |
Fam170a
|
family with sequence similarity 170, member A |
| chr2_-_120439981 | 4.84 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr17_-_29768586 | 4.82 |
ENSMUST00000234305.2
ENSMUST00000234648.2 ENSMUST00000234979.2 |
Gm17657
Tmem217
|
predicted gene, 17657 transmembrane protein 217 |
| chr10_-_38998272 | 4.81 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr6_+_41248311 | 4.79 |
ENSMUST00000103281.3
|
Trbv29
|
T cell receptor beta, variable 29 |
| chr14_-_87378641 | 4.78 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr17_-_71833752 | 4.67 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chr14_+_43781033 | 4.66 |
ENSMUST00000169023.2
|
Gm5799
|
predicted gene 5799 |
| chr11_-_103529678 | 4.65 |
ENSMUST00000107014.8
ENSMUST00000021328.8 |
Lyzl6
|
lysozyme-like 6 |
| chr11_-_78056347 | 4.64 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr17_+_37269468 | 4.61 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr6_-_149090146 | 4.61 |
ENSMUST00000095319.10
ENSMUST00000141346.2 ENSMUST00000111535.8 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr16_+_32066037 | 4.57 |
ENSMUST00000141820.8
ENSMUST00000178573.8 ENSMUST00000023474.4 ENSMUST00000135289.2 |
Wdr53
|
WD repeat domain 53 |
| chr7_-_101859308 | 4.57 |
ENSMUST00000070165.7
ENSMUST00000211235.2 ENSMUST00000211022.2 |
Nup98
|
nucleoporin 98 |
| chr13_+_22129246 | 4.57 |
ENSMUST00000176511.8
ENSMUST00000102978.8 ENSMUST00000152258.9 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr11_+_62842019 | 4.56 |
ENSMUST00000035854.4
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr17_-_56607250 | 4.56 |
ENSMUST00000233911.2
|
Arrdc5
|
arrestin domain containing 5 |
| chr5_-_82272549 | 4.54 |
ENSMUST00000188072.2
ENSMUST00000185410.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr4_-_86476502 | 4.52 |
ENSMUST00000030216.6
|
Saxo1
|
stabilizer of axonemal microtubules 1 |
| chr8_+_12623016 | 4.52 |
ENSMUST00000210276.2
ENSMUST00000010579.8 ENSMUST00000209428.2 |
Spaca7
|
sperm acrosome associated 7 |
| chr4_+_111577382 | 4.48 |
ENSMUST00000084354.4
|
Spata6
|
spermatogenesis associated 6 |
| chr14_+_43985323 | 4.48 |
ENSMUST00000100643.10
ENSMUST00000179860.2 |
Gm6526
|
predicted gene 6526 |
| chr3_-_140918116 | 4.48 |
ENSMUST00000057860.3
|
Pdha2
|
pyruvate dehydrogenase E1 alpha 2 |
| chr14_-_55108384 | 4.44 |
ENSMUST00000146642.2
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr7_+_46700349 | 4.41 |
ENSMUST00000010451.8
|
Tmem86a
|
transmembrane protein 86A |
| chr4_+_32238950 | 4.41 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr17_+_35201130 | 4.37 |
ENSMUST00000173004.2
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_-_52590686 | 4.35 |
ENSMUST00000098768.3
ENSMUST00000213843.2 |
AI593442
|
expressed sequence AI593442 |
| chrX_+_117336912 | 4.35 |
ENSMUST00000072518.8
|
Tgif2lx2
|
TGFB-induced factor homeobox 2-like, X-linked 2 |
| chr3_+_153549846 | 4.31 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr16_+_34470290 | 4.26 |
ENSMUST00000148562.8
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr2_+_155907100 | 4.25 |
ENSMUST00000038860.12
|
Spag4
|
sperm associated antigen 4 |
| chr18_+_35987791 | 4.20 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr3_+_40754448 | 4.20 |
ENSMUST00000026858.11
|
Plk4
|
polo like kinase 4 |
| chr11_-_70128587 | 4.20 |
ENSMUST00000108576.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr7_-_118183892 | 4.17 |
ENSMUST00000044195.6
|
Tmc7
|
transmembrane channel-like gene family 7 |
| chr4_+_42438970 | 4.15 |
ENSMUST00000238328.2
|
Gm21586
|
predicted gene, 21586 |
| chr18_+_14916295 | 4.14 |
ENSMUST00000234789.2
ENSMUST00000169862.2 |
Taf4b
|
TATA-box binding protein associated factor 4b |
| chr11_-_69871320 | 4.11 |
ENSMUST00000143175.2
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr14_+_70815250 | 4.10 |
ENSMUST00000228554.2
|
Nudt18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr2_-_124965537 | 4.07 |
ENSMUST00000142718.8
ENSMUST00000152367.8 ENSMUST00000067780.10 ENSMUST00000147105.8 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr2_-_153712996 | 4.01 |
ENSMUST00000028982.5
|
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr11_+_79230618 | 4.01 |
ENSMUST00000219057.2
ENSMUST00000108251.9 ENSMUST00000071325.9 |
Nf1
|
neurofibromin 1 |
| chr4_+_42293921 | 3.92 |
ENSMUST00000238203.2
|
Gm21953
|
predicted gene, 21953 |
| chr11_-_8989582 | 3.92 |
ENSMUST00000043377.6
|
Sun3
|
Sad1 and UNC84 domain containing 3 |
| chr4_-_123644091 | 3.91 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr10_+_80465481 | 3.91 |
ENSMUST00000085435.7
|
Csnk1g2
|
casein kinase 1, gamma 2 |
| chr4_+_100336003 | 3.91 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr13_-_111626562 | 3.85 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr5_+_101912939 | 3.81 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr14_-_64322849 | 3.81 |
ENSMUST00000089338.6
ENSMUST00000171503.8 |
Prss55
|
protease, serine 55 |
| chr14_+_96118660 | 3.79 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr11_-_77784922 | 3.79 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr2_+_109111083 | 3.75 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr7_-_112987770 | 3.75 |
ENSMUST00000079793.7
|
Pth
|
parathyroid hormone |
| chr3_+_28946760 | 3.74 |
ENSMUST00000099170.2
|
Gm1527
|
predicted gene 1527 |
| chr4_+_32238712 | 3.73 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chrX_+_11165496 | 3.69 |
ENSMUST00000188439.2
|
H2al1a
|
H2A histone family member L1A |
| chr6_+_136805808 | 3.68 |
ENSMUST00000052702.7
|
BC049715
|
cDNA sequence BC049715 |
| chr9_-_59393893 | 3.66 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr2_-_120439826 | 3.65 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr14_+_33774640 | 3.61 |
ENSMUST00000226211.2
|
Antxrl
|
anthrax toxin receptor-like |
| chr5_+_33978035 | 3.60 |
ENSMUST00000075812.11
ENSMUST00000114397.9 ENSMUST00000155880.8 |
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr17_+_35200823 | 3.60 |
ENSMUST00000114011.11
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_24211879 | 3.58 |
ENSMUST00000234781.2
|
Gm50092
|
predicted gene, 50092 |
| chr2_+_148640705 | 3.57 |
ENSMUST00000028931.10
ENSMUST00000109947.2 |
Cst8
|
cystatin 8 (cystatin-related epididymal spermatogenic) |
| chr3_+_127347099 | 3.56 |
ENSMUST00000043108.9
ENSMUST00000196141.5 ENSMUST00000195955.2 |
Zgrf1
|
zinc finger, GRF-type containing 1 |
| chr11_-_118233326 | 3.54 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr14_-_21792938 | 3.52 |
ENSMUST00000120956.8
|
Dusp13
|
dual specificity phosphatase 13 |
| chr4_-_41774097 | 3.52 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr11_+_115353290 | 3.51 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr11_-_69214593 | 3.50 |
ENSMUST00000092973.6
|
Cntrob
|
centrobin, centrosomal BRCA2 interacting protein |
| chr7_+_63094489 | 3.48 |
ENSMUST00000058476.14
|
Otud7a
|
OTU domain containing 7A |
| chr3_+_125197722 | 3.48 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr14_+_75479727 | 3.47 |
ENSMUST00000022576.10
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr19_+_6135013 | 3.46 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr17_-_32255287 | 3.43 |
ENSMUST00000238192.2
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr10_-_128383508 | 3.39 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr6_+_129510117 | 3.37 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr11_+_82782938 | 3.31 |
ENSMUST00000018988.6
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr4_+_41942037 | 3.30 |
ENSMUST00000181518.2
|
Gm20878
|
predicted gene, 20878 |
| chr14_+_33775115 | 3.27 |
ENSMUST00000227979.2
|
Antxrl
|
anthrax toxin receptor-like |
| chr10_-_82152373 | 3.27 |
ENSMUST00000217661.2
|
4932415D10Rik
|
RIKEN cDNA 4932415D10 gene |
| chr7_+_28455563 | 3.24 |
ENSMUST00000178767.3
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr5_+_24369961 | 3.18 |
ENSMUST00000049887.13
|
Nupl2
|
nucleoporin like 2 |
| chr7_-_109092834 | 3.16 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chrX_+_30768610 | 3.08 |
ENSMUST00000179532.2
|
Btbd35f29
|
BTB domain containing 35, family member 29 |
| chr8_-_61407760 | 3.04 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr7_-_45375205 | 3.03 |
ENSMUST00000094424.7
|
Spaca4
|
sperm acrosome associated 4 |
| chr2_-_164080358 | 3.03 |
ENSMUST00000044953.3
|
Svs2
|
seminal vesicle secretory protein 2 |
| chr18_-_3309858 | 3.02 |
ENSMUST00000144496.8
ENSMUST00000154715.8 |
Crem
|
cAMP responsive element modulator |
| chr17_+_36290743 | 3.01 |
ENSMUST00000087200.4
|
Gnl1
|
guanine nucleotide binding protein-like 1 |
| chr11_-_51153870 | 3.01 |
ENSMUST00000054226.3
|
BC049762
|
cDNA sequence BC049762 |
| chr3_+_90011435 | 3.01 |
ENSMUST00000029548.9
ENSMUST00000200410.2 |
Nup210l
|
nucleoporin 210-like |
| chr8_-_56359983 | 3.00 |
ENSMUST00000053441.5
|
Adam29
|
a disintegrin and metallopeptidase domain 29 |
| chr6_+_41092928 | 2.98 |
ENSMUST00000194399.2
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
| chr7_+_140641010 | 2.95 |
ENSMUST00000048002.7
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr14_-_44962444 | 2.94 |
ENSMUST00000165003.8
ENSMUST00000169062.2 |
Gm8267
|
predicted gene 8267 |
| chrX_+_9216866 | 2.93 |
ENSMUST00000178196.3
|
H2al1k
|
H2A histone family member L1K |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 19.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 2.0 | 9.9 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 2.0 | 7.8 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 1.8 | 12.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.8 | 5.3 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.7 | 32.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 1.4 | 4.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 1.2 | 10.7 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 1.2 | 9.4 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 1.2 | 3.5 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.2 | 6.9 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 1.1 | 10.6 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 1.0 | 5.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.0 | 5.9 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.9 | 3.7 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.9 | 10.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.9 | 2.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.9 | 11.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.9 | 3.6 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.9 | 2.6 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.8 | 2.5 | GO:0097065 | cervix development(GO:0060067) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.8 | 8.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.8 | 12.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.8 | 2.4 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.7 | 2.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.7 | 3.5 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.7 | 4.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.6 | 6.5 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.6 | 11.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.6 | 7.5 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.6 | 1.9 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.6 | 5.6 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.6 | 8.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.6 | 1.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.6 | 1.8 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.6 | 3.6 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.6 | 3.5 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.6 | 4.7 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.6 | 2.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.6 | 1.7 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.6 | 6.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.5 | 2.2 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.5 | 2.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.5 | 4.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.5 | 3.5 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.5 | 4.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.5 | 1.9 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.5 | 2.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.5 | 1.4 | GO:1901738 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin A metabolic process(GO:1901738) |
| 0.4 | 7.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.4 | 1.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.4 | 7.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 2.2 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.4 | 2.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.4 | 2.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.4 | 4.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 1.6 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.4 | 2.0 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.4 | 14.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.4 | 2.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 1.1 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.4 | 2.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.4 | 7.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 1.4 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.3 | 4.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 25.3 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.3 | 1.0 | GO:0006233 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.3 | 1.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 5.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 1.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 2.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.3 | 2.5 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.3 | 2.8 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 1.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 3.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.3 | 3.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 10.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 2.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 13.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.3 | 0.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.3 | 2.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 1.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.3 | 1.4 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.3 | 1.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.3 | 2.1 | GO:1901750 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 2.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 0.8 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.3 | 30.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.3 | 3.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 0.7 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.2 | 1.5 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.2 | 0.9 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 1.4 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.2 | 4.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 5.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 4.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.8 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 7.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 1.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.2 | 2.9 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.6 | GO:1902524 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 2.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 27.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 0.7 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 1.7 | GO:0071684 | blastocyst hatching(GO:0001835) DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 1.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 3.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.2 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 4.9 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.2 | 5.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 1.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 2.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 1.3 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.9 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 3.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 2.6 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.1 | 0.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 2.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.6 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 6.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 1.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 2.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.9 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 1.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.5 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 7.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 3.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 5.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.6 | GO:0060770 | regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 1.8 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 2.0 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 1.4 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.1 | 4.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 5.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 22.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 6.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 1.2 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.1 | 9.4 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.7 | GO:0032202 | protection from non-homologous end joining at telomere(GO:0031848) telomere assembly(GO:0032202) |
| 0.1 | 0.3 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.1 | 0.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 3.4 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.1 | 0.9 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 1.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 1.0 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.5 | GO:1903333 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 3.0 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 1.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.8 | GO:2000394 | astrocyte cell migration(GO:0043615) regulation of GTP binding(GO:1904424) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 0.4 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 4.1 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.6 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 2.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 3.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 2.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 6.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.1 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.9 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 1.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.3 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 6.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.9 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 8.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 3.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 1.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0061205 | paramesonephric duct development(GO:0061205) |
| 0.0 | 0.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.7 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.2 | GO:0051028 | mRNA transport(GO:0051028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 2.0 | 9.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 1.5 | 4.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 1.3 | 9.4 | GO:0098536 | deuterosome(GO:0098536) |
| 1.3 | 17.9 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 1.2 | 3.6 | GO:0034359 | mature chylomicron(GO:0034359) |
| 1.1 | 4.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.1 | 5.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 1.1 | 4.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 1.0 | 28.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.0 | 2.9 | GO:1990427 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 0.9 | 8.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.8 | 11.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.8 | 11.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.7 | 13.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.7 | 9.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 4.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.6 | 4.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.6 | 4.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.5 | 1.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 7.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 3.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 11.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 2.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.4 | 3.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 3.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 4.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.4 | 2.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 5.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 6.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 1.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 43.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 2.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 3.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 1.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 11.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 2.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 2.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.2 | 9.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 19.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 0.8 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 0.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 4.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 5.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.6 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 15.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.4 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 1.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 5.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.6 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 1.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 3.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 4.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 13.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 2.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 1.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 36.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 2.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 2.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 5.3 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 3.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 5.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 3.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 23.0 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 3.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 5.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 1.5 | 10.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.5 | 6.0 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 1.5 | 4.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.4 | 13.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.4 | 4.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 1.3 | 3.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.1 | 12.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.1 | 11.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.1 | 5.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 1.1 | 5.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.0 | 7.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.8 | 2.5 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.7 | 2.2 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.7 | 2.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.7 | 5.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.7 | 12.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.7 | 12.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.6 | 4.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 3.8 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.6 | 2.5 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.6 | 9.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.6 | 2.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.5 | 2.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 1.6 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.5 | 1.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.5 | 12.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.5 | 2.8 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.5 | 1.9 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.5 | 4.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.4 | 1.8 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.4 | 11.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 1.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 12.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 6.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.4 | 3.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.4 | 1.1 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.4 | 3.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 1.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 4.1 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 2.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 1.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 1.0 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.3 | 1.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 4.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 0.9 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.3 | 8.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 3.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 0.5 | GO:0032564 | dATP binding(GO:0032564) |
| 0.3 | 3.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.7 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-3 binding(GO:0019978) |
| 0.2 | 7.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 1.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.4 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 0.7 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.2 | 1.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 12.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 7.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 2.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 8.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 2.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 3.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 1.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 12.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 12.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.2 | 0.6 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 4.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.4 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 3.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 34.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 1.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 3.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 26.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 5.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 1.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.9 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 1.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 4.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 11.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.2 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 3.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 1.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 4.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 2.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 4.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.4 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 3.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 4.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 5.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 7.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 6.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 6.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 14.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 14.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 25.1 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 1.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 18.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 1.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 9.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 6.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 3.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 8.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 5.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.3 | 8.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 6.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 16.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 2.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 2.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 5.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 6.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 3.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 5.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 5.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 2.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 5.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 7.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 2.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 8.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |