Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
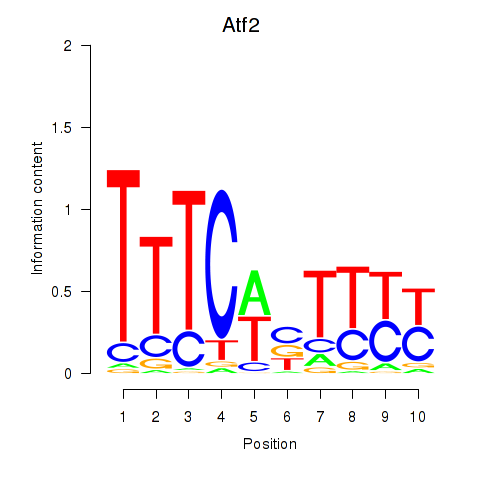
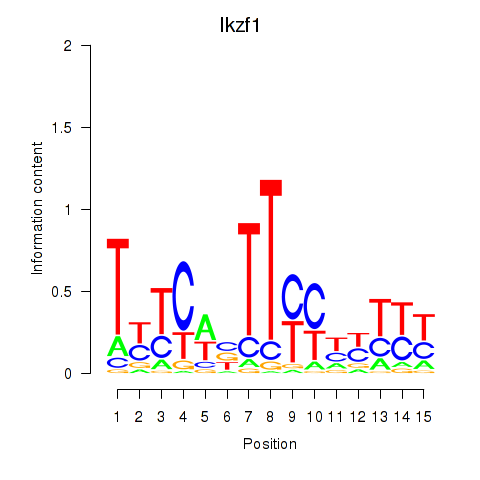
Results for Atf2_Ikzf1
Z-value: 2.37
Transcription factors associated with Atf2_Ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf2
|
ENSMUSG00000027104.19 | Atf2 |
|
Ikzf1
|
ENSMUSG00000018654.18 | Ikzf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf2 | mm39_v1_chr2_-_73722932_73722983 | 0.41 | 3.9e-04 | Click! |
| Ikzf1 | mm39_v1_chr11_+_11635908_11635949 | -0.12 | 3.2e-01 | Click! |
Activity profile of Atf2_Ikzf1 motif
Sorted Z-values of Atf2_Ikzf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf2_Ikzf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_119978773 | 22.90 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr1_-_169575203 | 22.79 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chrX_+_165127688 | 22.40 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr2_-_180798785 | 22.34 |
ENSMUST00000055990.8
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr12_-_90705212 | 21.72 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr10_+_29019645 | 20.72 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr3_-_79645101 | 20.63 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr3_+_8574420 | 20.41 |
ENSMUST00000029002.9
|
Stmn2
|
stathmin-like 2 |
| chr9_-_117081518 | 20.39 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr13_-_78347876 | 19.38 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr6_+_141470105 | 19.06 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr13_+_42862957 | 17.00 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr1_+_66214445 | 16.89 |
ENSMUST00000114017.8
ENSMUST00000114015.8 |
Map2
|
microtubule-associated protein 2 |
| chr9_+_27702243 | 16.67 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr3_-_116047148 | 16.27 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr1_+_9671388 | 16.12 |
ENSMUST00000088666.4
|
Vxn
|
vexin |
| chr1_-_173161069 | 14.53 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr1_+_158189831 | 13.67 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr11_+_100902572 | 13.36 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr17_-_90763300 | 13.35 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr2_-_57942844 | 13.32 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr5_-_103359117 | 13.21 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr13_-_78345932 | 12.98 |
ENSMUST00000127137.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr13_+_105580147 | 12.96 |
ENSMUST00000022235.6
|
Htr1a
|
5-hydroxytryptamine (serotonin) receptor 1A |
| chr5_-_70999547 | 12.88 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chrX_-_87159237 | 12.87 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr8_+_40807344 | 12.87 |
ENSMUST00000136835.2
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chrX_+_135723420 | 12.73 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr8_+_57908920 | 12.68 |
ENSMUST00000034023.4
|
Scrg1
|
scrapie responsive gene 1 |
| chr12_+_52746158 | 12.59 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr13_+_54519161 | 12.58 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chrX_+_135723531 | 12.33 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chrX_+_40490353 | 11.93 |
ENSMUST00000165288.2
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr11_-_42072990 | 11.73 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr2_-_167030706 | 11.68 |
ENSMUST00000207917.2
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chrX_+_135893894 | 11.13 |
ENSMUST00000113067.8
ENSMUST00000101227.3 |
Zcchc18
|
zinc finger, CCHC domain containing 18 |
| chr2_-_129541753 | 11.09 |
ENSMUST00000028883.12
|
Pdyn
|
prodynorphin |
| chr9_-_117081185 | 11.03 |
ENSMUST00000111772.10
ENSMUST00000174868.8 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr9_-_91247809 | 10.95 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr5_+_37332834 | 10.90 |
ENSMUST00000208827.2
ENSMUST00000207619.2 |
Gm1043
|
predicted gene 1043 |
| chr15_-_37792237 | 10.90 |
ENSMUST00000168992.8
ENSMUST00000148652.9 |
Ncald
|
neurocalcin delta |
| chr2_+_170573727 | 10.83 |
ENSMUST00000029075.5
|
Dok5
|
docking protein 5 |
| chr15_+_82159094 | 10.58 |
ENSMUST00000116423.3
ENSMUST00000230418.2 |
Septin3
|
septin 3 |
| chr6_+_141470261 | 10.45 |
ENSMUST00000203140.2
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr11_-_42072920 | 10.41 |
ENSMUST00000207274.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr1_+_158190090 | 10.23 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr16_+_7042168 | 10.19 |
ENSMUST00000231088.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr16_+_96268806 | 10.18 |
ENSMUST00000061739.9
|
Pcp4
|
Purkinje cell protein 4 |
| chr14_+_55173696 | 10.04 |
ENSMUST00000037814.8
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chrX_-_94209913 | 9.86 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr16_+_41353360 | 9.79 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr18_-_31450095 | 9.61 |
ENSMUST00000139924.2
ENSMUST00000153060.8 |
Rit2
|
Ras-like without CAAX 2 |
| chr3_+_156267429 | 9.54 |
ENSMUST00000074015.11
|
Negr1
|
neuronal growth regulator 1 |
| chr11_-_99313078 | 9.53 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr11_+_31950452 | 9.50 |
ENSMUST00000109409.8
ENSMUST00000020537.9 |
Nsg2
|
neuron specific gene family member 2 |
| chr11_+_87651359 | 9.40 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr14_-_88708782 | 9.36 |
ENSMUST00000192557.2
ENSMUST00000061628.7 |
Pcdh20
|
protocadherin 20 |
| chr2_+_136555364 | 9.35 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr1_-_46927230 | 9.30 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_+_135894030 | 9.16 |
ENSMUST00000033804.5
|
Zcchc18
|
zinc finger, CCHC domain containing 18 |
| chr2_+_97298002 | 9.14 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr1_+_66214431 | 9.11 |
ENSMUST00000156636.9
|
Map2
|
microtubule-associated protein 2 |
| chr6_+_96090127 | 9.08 |
ENSMUST00000122120.8
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr2_+_151923449 | 9.04 |
ENSMUST00000064061.4
|
Scrt2
|
scratch family zinc finger 2 |
| chr2_+_67578556 | 8.91 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr2_+_121188195 | 8.82 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chrX_+_134643435 | 8.80 |
ENSMUST00000096321.3
ENSMUST00000113144.8 ENSMUST00000113147.8 ENSMUST00000113145.8 |
Armcx5
Gprasp1
|
armadillo repeat containing, X-linked 5 G protein-coupled receptor associated sorting protein 1 |
| chr1_+_66426127 | 8.77 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr18_+_37622518 | 8.77 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr6_-_25689781 | 8.58 |
ENSMUST00000200812.2
|
Gpr37
|
G protein-coupled receptor 37 |
| chr2_-_17465410 | 8.51 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr11_-_69496655 | 8.50 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr1_+_153541412 | 8.50 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr10_+_85222677 | 8.49 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr5_+_71857261 | 8.43 |
ENSMUST00000031122.9
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr16_+_41353212 | 8.28 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr9_-_75591274 | 8.26 |
ENSMUST00000214244.2
ENSMUST00000213324.2 ENSMUST00000034699.8 |
Scg3
|
secretogranin III |
| chr15_-_98851566 | 8.26 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr5_-_104261285 | 8.09 |
ENSMUST00000199947.2
|
Sparcl1
|
SPARC-like 1 |
| chr1_-_38704028 | 8.03 |
ENSMUST00000039827.14
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr14_-_67106037 | 7.91 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr2_-_66240408 | 7.86 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr13_+_46571910 | 7.85 |
ENSMUST00000037923.5
|
Rbm24
|
RNA binding motif protein 24 |
| chr3_-_87170903 | 7.83 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr11_+_69917640 | 7.83 |
ENSMUST00000135916.9
ENSMUST00000232659.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_+_158189900 | 7.79 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chrX_-_133012600 | 7.68 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr11_-_83959999 | 7.37 |
ENSMUST00000138208.2
|
Dusp14
|
dual specificity phosphatase 14 |
| chr1_-_55265925 | 7.35 |
ENSMUST00000027121.15
ENSMUST00000114428.3 |
Rftn2
|
raftlin family member 2 |
| chrX_-_94701983 | 7.31 |
ENSMUST00000119640.8
ENSMUST00000120620.8 ENSMUST00000044382.7 |
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr15_+_82159398 | 7.29 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chr17_-_37334240 | 7.26 |
ENSMUST00000102665.11
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr9_+_37278647 | 7.25 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr1_-_174749379 | 7.23 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr12_-_79027531 | 7.19 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr16_-_34334314 | 7.17 |
ENSMUST00000151491.8
ENSMUST00000114960.9 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr3_-_72964276 | 7.16 |
ENSMUST00000192477.2
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr13_+_41071077 | 7.14 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr5_+_137551774 | 7.10 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr2_-_6889783 | 7.05 |
ENSMUST00000170438.8
ENSMUST00000114924.10 ENSMUST00000114934.11 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr4_-_110144676 | 7.03 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr14_+_55173936 | 7.00 |
ENSMUST00000227441.2
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr9_-_91247831 | 6.84 |
ENSMUST00000065360.5
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr11_+_89921121 | 6.84 |
ENSMUST00000092788.4
|
Tmem100
|
transmembrane protein 100 |
| chr2_+_14878480 | 6.83 |
ENSMUST00000114719.7
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_-_110148081 | 6.81 |
ENSMUST00000142722.2
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr2_+_49509288 | 6.80 |
ENSMUST00000028102.14
|
Kif5c
|
kinesin family member 5C |
| chr1_+_75351914 | 6.72 |
ENSMUST00000087122.12
|
Speg
|
SPEG complex locus |
| chr10_-_102326286 | 6.69 |
ENSMUST00000020040.5
|
Nts
|
neurotensin |
| chrX_+_82898974 | 6.68 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chr9_+_104443876 | 6.67 |
ENSMUST00000157006.8
|
Cpne4
|
copine IV |
| chr3_-_87171005 | 6.66 |
ENSMUST00000146512.2
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr7_+_45349267 | 6.66 |
ENSMUST00000003360.10
|
Car11
|
carbonic anhydrase 11 |
| chr11_+_31950496 | 6.63 |
ENSMUST00000093219.4
|
Nsg2
|
neuron specific gene family member 2 |
| chr12_+_88689638 | 6.57 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr18_+_23548192 | 6.57 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr15_-_79718423 | 6.55 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr2_-_6889685 | 6.53 |
ENSMUST00000183091.8
ENSMUST00000182851.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr2_-_115895528 | 6.53 |
ENSMUST00000028639.13
ENSMUST00000102538.11 |
Meis2
|
Meis homeobox 2 |
| chr10_-_20600797 | 6.52 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr14_-_124914516 | 6.44 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr12_-_4891435 | 6.42 |
ENSMUST00000219880.2
ENSMUST00000020964.7 |
Fkbp1b
|
FK506 binding protein 1b |
| chr5_+_57875309 | 6.42 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr7_-_6733411 | 6.36 |
ENSMUST00000239104.2
ENSMUST00000051209.11 |
Peg3
|
paternally expressed 3 |
| chr1_+_171077984 | 6.35 |
ENSMUST00000111315.9
ENSMUST00000219033.2 |
Adamts4
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 4 |
| chr2_-_131953359 | 6.34 |
ENSMUST00000128899.2
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chrX_+_149981074 | 6.29 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr6_-_28134544 | 6.28 |
ENSMUST00000115323.8
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr1_+_140173787 | 6.21 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr18_+_37085673 | 6.20 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chrX_+_92718695 | 6.20 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr9_+_36743980 | 6.18 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_+_96295011 | 6.15 |
ENSMUST00000233816.2
|
Pcp4
|
Purkinje cell protein 4 |
| chr2_+_157401998 | 6.08 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr12_+_83572774 | 5.99 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr18_+_59195534 | 5.95 |
ENSMUST00000058633.9
ENSMUST00000175897.8 ENSMUST00000118510.8 ENSMUST00000175830.2 |
Minar2
|
membrane integral NOTCH2 associated receptor 2 |
| chr17_+_43700327 | 5.89 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr10_+_86599836 | 5.86 |
ENSMUST00000218802.2
|
Gm49358
|
predicted gene, 49358 |
| chr18_+_23548534 | 5.83 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr2_+_65676111 | 5.82 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr9_+_50664288 | 5.78 |
ENSMUST00000214962.2
ENSMUST00000216755.2 |
Cryab
|
crystallin, alpha B |
| chr8_+_106075475 | 5.78 |
ENSMUST00000073149.7
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5 |
| chr6_-_23839136 | 5.77 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr14_-_55150547 | 5.66 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr8_-_85500010 | 5.63 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr18_+_69726654 | 5.61 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr1_-_130656985 | 5.60 |
ENSMUST00000189534.7
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr3_-_50398027 | 5.59 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr10_+_90412432 | 5.56 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_-_91396154 | 5.54 |
ENSMUST00000161402.10
ENSMUST00000054059.15 ENSMUST00000072671.14 ENSMUST00000174331.8 |
Nrxn1
|
neurexin I |
| chr1_-_75240551 | 5.53 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr2_+_177783713 | 5.52 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr7_+_3381434 | 5.49 |
ENSMUST00000092891.6
|
Cacng7
|
calcium channel, voltage-dependent, gamma subunit 7 |
| chr1_-_22386016 | 5.44 |
ENSMUST00000164877.8
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chrX_+_139243012 | 5.44 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr1_-_154602102 | 5.42 |
ENSMUST00000187541.7
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr18_+_37610858 | 5.40 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr5_+_137551790 | 5.33 |
ENSMUST00000136565.8
ENSMUST00000149292.8 ENSMUST00000125489.2 |
Actl6b
|
actin-like 6B |
| chr16_-_45544960 | 5.33 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr2_-_180956293 | 5.28 |
ENSMUST00000103045.4
|
Stmn3
|
stathmin-like 3 |
| chr10_+_126914755 | 5.25 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_-_31133622 | 5.20 |
ENSMUST00000115230.2
ENSMUST00000130560.8 |
Apod
|
apolipoprotein D |
| chr9_-_50605079 | 5.19 |
ENSMUST00000120622.2
|
Dixdc1
|
DIX domain containing 1 |
| chr12_+_95658987 | 5.19 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr18_+_59195354 | 5.18 |
ENSMUST00000165666.9
|
Minar2
|
membrane integral NOTCH2 associated receptor 2 |
| chr7_-_81143631 | 5.18 |
ENSMUST00000082090.15
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr10_+_56255184 | 5.18 |
ENSMUST00000220069.2
|
Gja1
|
gap junction protein, alpha 1 |
| chrX_-_134968985 | 5.18 |
ENSMUST00000049130.8
|
Bex2
|
brain expressed X-linked 2 |
| chr1_-_65162267 | 5.13 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr12_+_61570669 | 5.13 |
ENSMUST00000055815.14
ENSMUST00000119481.2 |
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr7_+_130179063 | 5.11 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr2_+_158508609 | 5.09 |
ENSMUST00000103116.10
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr10_+_59942274 | 5.09 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr12_+_84498196 | 5.03 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr18_+_37474755 | 5.02 |
ENSMUST00000053037.5
|
Pcdhb7
|
protocadherin beta 7 |
| chr10_+_29087658 | 5.00 |
ENSMUST00000213489.2
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr18_+_66005891 | 4.99 |
ENSMUST00000173985.10
|
Grp
|
gastrin releasing peptide |
| chr9_-_77255069 | 4.99 |
ENSMUST00000184848.8
ENSMUST00000184415.8 |
Mlip
|
muscular LMNA-interacting protein |
| chrX_+_13499008 | 4.98 |
ENSMUST00000096492.4
|
Gpr34
|
G protein-coupled receptor 34 |
| chr12_-_76865148 | 4.95 |
ENSMUST00000118604.8
|
Rab15
|
RAB15, member RAS oncogene family |
| chr14_+_66581818 | 4.94 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chr17_-_37334091 | 4.93 |
ENSMUST00000167275.3
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr6_+_77219698 | 4.93 |
ENSMUST00000161677.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr2_+_14828903 | 4.90 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_+_22875494 | 4.88 |
ENSMUST00000090568.7
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
| chr4_-_41697040 | 4.88 |
ENSMUST00000102962.10
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr4_+_85123358 | 4.87 |
ENSMUST00000107188.10
|
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr3_-_19749489 | 4.84 |
ENSMUST00000061294.5
|
Crh
|
corticotropin releasing hormone |
| chr1_-_132318039 | 4.83 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr11_-_3864664 | 4.82 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr14_-_80008745 | 4.80 |
ENSMUST00000039568.11
ENSMUST00000195355.2 |
Pcdh8
|
protocadherin 8 |
| chr1_-_69147185 | 4.78 |
ENSMUST00000121473.8
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr11_+_69909659 | 4.78 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr9_-_54381409 | 4.71 |
ENSMUST00000127880.8
|
Dmxl2
|
Dmx-like 2 |
| chr2_-_26917921 | 4.70 |
ENSMUST00000102890.11
ENSMUST00000153388.2 ENSMUST00000045702.6 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr10_+_90412114 | 4.67 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_105398346 | 4.66 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr10_+_29087602 | 4.65 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr19_-_19088543 | 4.65 |
ENSMUST00000112832.8
|
Rorb
|
RAR-related orphan receptor beta |
| chr11_-_109188947 | 4.65 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 33.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 6.9 | 20.6 | GO:0060618 | nipple development(GO:0060618) |
| 5.1 | 20.4 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 3.3 | 26.8 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 3.2 | 22.7 | GO:0061743 | motor learning(GO:0061743) |
| 3.2 | 12.9 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 3.2 | 9.5 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 3.1 | 18.9 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 3.1 | 9.4 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 3.1 | 18.4 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 3.0 | 35.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 2.9 | 11.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 2.4 | 7.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 2.2 | 2.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 2.1 | 6.3 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 2.1 | 6.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 2.0 | 20.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.9 | 22.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 1.9 | 5.7 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.9 | 13.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.8 | 42.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.8 | 9.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.8 | 1.8 | GO:0072249 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.7 | 5.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.7 | 5.2 | GO:0010232 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 1.7 | 1.7 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 1.7 | 8.5 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.7 | 6.8 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.5 | 7.6 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 1.5 | 4.5 | GO:0003032 | detection of oxygen(GO:0003032) |
| 1.5 | 8.9 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 1.5 | 4.5 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.5 | 11.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 1.4 | 4.2 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.4 | 25.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.4 | 15.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.4 | 4.1 | GO:0061193 | taste bud development(GO:0061193) |
| 1.3 | 4.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.3 | 3.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 1.3 | 7.8 | GO:0035127 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.3 | 6.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.3 | 5.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.3 | 5.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 1.3 | 31.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 1.3 | 7.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 1.2 | 5.0 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 1.2 | 11.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.2 | 4.9 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 1.2 | 17.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 1.2 | 4.9 | GO:0003360 | brainstem development(GO:0003360) |
| 1.2 | 34.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.2 | 17.0 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.2 | 4.8 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 1.2 | 20.3 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 1.2 | 6.0 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.2 | 7.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 1.2 | 3.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 1.2 | 4.7 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 1.2 | 3.5 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.2 | 12.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 1.1 | 13.6 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.0 | 5.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 1.0 | 3.0 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.0 | 3.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 1.0 | 5.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.0 | 3.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.9 | 11.3 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.9 | 11.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.9 | 10.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.9 | 9.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.9 | 4.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.9 | 2.8 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.9 | 8.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.9 | 8.3 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.9 | 12.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.9 | 2.7 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.9 | 4.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.9 | 2.6 | GO:0097688 | glutamate receptor clustering(GO:0097688) |
| 0.9 | 9.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.9 | 8.6 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.9 | 1.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.9 | 15.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.8 | 26.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.8 | 41.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.8 | 2.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.8 | 6.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.8 | 4.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.8 | 0.8 | GO:0050883 | vestibular nucleus development(GO:0021750) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.8 | 1.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.8 | 3.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.8 | 3.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.8 | 7.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.8 | 2.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.8 | 4.6 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.8 | 5.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.7 | 3.7 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.7 | 3.7 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.7 | 4.4 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.7 | 2.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.7 | 2.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.7 | 10.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.7 | 2.9 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.7 | 8.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.7 | 12.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.7 | 5.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.7 | 2.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.7 | 2.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.7 | 2.8 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.7 | 33.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.6 | 23.3 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.6 | 9.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.6 | 14.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.6 | 1.9 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.6 | 3.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.6 | 1.9 | GO:0071317 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.6 | 2.5 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.6 | 4.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.6 | 2.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.6 | 3.0 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.6 | 5.3 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.6 | 3.5 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.6 | 3.4 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.6 | 7.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 3.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.6 | 3.3 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.5 | 1.6 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.5 | 18.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 6.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 6.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 | 5.7 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.5 | 1.6 | GO:1900063 | mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.5 | 1.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.5 | 8.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.5 | 7.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 1.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.5 | 3.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 3.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.5 | 2.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.5 | 14.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 4.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.5 | 1.9 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.5 | 2.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.5 | 16.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 1.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.4 | 1.7 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.4 | 3.5 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.4 | 5.6 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.4 | 2.6 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 | 24.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.4 | 3.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.4 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.4 | 11.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.4 | 15.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 5.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.4 | 1.6 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.4 | 10.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 1.9 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.4 | 4.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 1.2 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.4 | 2.3 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.4 | 1.5 | GO:0060733 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.4 | 1.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.4 | 2.6 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.4 | 7.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 1.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 2.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 5.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 2.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.4 | 3.6 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.4 | 4.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.4 | 4.7 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 4.7 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.4 | 1.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.4 | 1.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.4 | 5.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.4 | 8.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 1.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.4 | 1.8 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.3 | 1.4 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.3 | 3.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.3 | 1.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 4.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 2.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.3 | 1.4 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 | 2.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.3 | 3.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 13.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 5.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 1.0 | GO:0036233 | regulation of amino acid import(GO:0010958) glycine import(GO:0036233) |
| 0.3 | 5.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 2.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.3 | 5.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 2.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.3 | 19.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 3.7 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 2.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 1.2 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.3 | 0.6 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.3 | 2.6 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.3 | 0.6 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.3 | 1.5 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.3 | 2.0 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.3 | 1.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 0.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 2.8 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.3 | 34.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 3.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 20.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.3 | 1.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 4.9 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.3 | 10.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.3 | 1.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.3 | 5.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.3 | 9.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 2.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.3 | 3.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 4.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.3 | 1.8 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.3 | 0.8 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.3 | 2.0 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.3 | 0.8 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.3 | 0.5 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.3 | 2.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 19.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 2.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 | 1.2 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.2 | 4.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.2 | 5.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 12.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.2 | 0.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 11.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 4.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 3.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.5 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.2 | 0.9 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 3.5 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.2 | 3.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 3.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 5.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 1.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 0.9 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 9.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 2.0 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.2 | 30.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.2 | 3.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 4.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 1.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 3.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 1.3 | GO:0045650 | regulation of macrophage differentiation(GO:0045649) negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 2.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.2 | 3.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 2.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 4.1 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 2.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 2.9 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 1.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 0.4 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.2 | 1.0 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 0.6 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.2 | 1.4 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 17.6 | GO:0007612 | learning(GO:0007612) |
| 0.2 | 0.8 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 9.1 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.2 | 1.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 2.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 3.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 1.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.2 | 1.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.2 | GO:0003163 | sinoatrial node development(GO:0003163) |
| 0.2 | 2.9 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 0.8 | GO:0060013 | righting reflex(GO:0060013) negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 2.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 0.9 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.2 | 0.7 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 3.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 3.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 2.9 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.2 | 1.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 1.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 1.1 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 11.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 0.5 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 6.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 0.9 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.2 | 5.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 2.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 1.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 4.0 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 1.6 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.2 | 0.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 1.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 | 5.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 | 1.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 1.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.2 | 0.9 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 3.7 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.4 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.4 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.1 | 2.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.6 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 3.2 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.6 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 2.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.1 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 4.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.4 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.4 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.5 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 1.6 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 2.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 2.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 6.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 2.7 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 15.5 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 1.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.5 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.4 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 14.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.5 | GO:0009227 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.1 | 0.7 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 3.9 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 1.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 4.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 4.7 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.2 | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.1 | 1.0 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.4 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 2.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 5.0 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.1 | 5.5 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 2.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 5.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 3.9 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 10.4 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 2.3 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 0.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 2.0 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 0.6 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 8.0 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 7.3 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.1 | 1.1 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.8 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.5 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 1.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 2.0 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 1.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.5 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 1.4 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 1.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 2.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 3.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 6.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 2.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 15.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 2.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.1 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.0 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.1 | 0.8 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.6 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 0.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 5.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 4.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 8.1 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 8.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.1 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.1 | 1.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 1.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 2.0 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 2.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 3.5 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.1 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.4 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 6.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.8 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 2.4 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 1.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 1.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.0 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.3 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 1.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0033026 | positive regulation of mast cell cytokine production(GO:0032765) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.0 | 0.7 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 5.4 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.3 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 4.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 1.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 1.7 | GO:0008366 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.6 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 1.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0071545 | inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 1.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.2 | GO:0090197 | positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.6 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0002067 | glandular epithelial cell differentiation(GO:0002067) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 1.8 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.4 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 4.5 | 40.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 4.4 | 21.9 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 3.7 | 14.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 3.2 | 22.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 2.4 | 9.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 2.2 | 8.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.9 | 7.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.8 | 10.7 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.8 | 14.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.7 | 13.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.6 | 4.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.5 | 42.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.3 | 24.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.3 | 7.8 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.2 | 3.5 | GO:0060187 | cell pole(GO:0060187) |
| 1.1 | 16.0 | GO:0005883 | neurofilament(GO:0005883) |
| 1.1 | 4.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.1 | 4.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.0 | 22.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.0 | 5.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 1.0 | 5.9 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 1.0 | 4.9 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 1.0 | 7.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.9 | 17.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.9 | 14.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.8 | 13.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.8 | 12.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.8 | 13.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 5.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.8 | 7.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.7 | 2.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.7 | 2.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.7 | 4.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.7 | 5.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.6 | 3.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.6 | 3.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.6 | 4.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.6 | 21.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.5 | 9.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 14.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.5 | 3.7 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.5 | 23.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.5 | 3.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.4 | 4.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 0.4 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.4 | 5.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.4 | 1.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 6.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 2.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 8.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 9.7 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.4 | 19.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 4.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 10.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 3.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.4 | 138.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.4 | 4.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.4 | 10.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 8.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 101.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 3.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 5.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 2.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 1.0 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.3 | 4.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 18.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 2.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 0.9 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.3 | 9.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 49.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.3 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 3.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 1.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 2.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 1.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 1.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 10.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 5.9 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 16.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 46.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 6.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 4.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 1.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 8.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 1.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 0.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 0.6 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.2 | 3.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 2.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 3.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 2.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 8.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 2.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.0 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 1.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 27.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 4.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 9.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 2.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.7 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 5.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 8.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.5 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 5.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 17.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.6 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 32.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 10.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 14.8 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 6.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 4.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 7.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 24.5 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 7.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 5.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 33.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 21.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 4.5 | 40.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 3.7 | 29.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 3.2 | 48.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.1 | 30.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 2.6 | 13.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.5 | 7.5 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 2.2 | 31.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.2 | 8.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 2.1 | 8.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 2.1 | 6.3 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 2.1 | 6.3 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 2.0 | 14.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.9 | 15.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.8 | 7.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.7 | 12.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.7 | 28.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.6 | 1.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 1.6 | 6.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.4 | 11.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.4 | 4.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.3 | 16.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.3 | 7.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.3 | 7.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 1.3 | 3.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.3 | 8.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.2 | 4.8 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 1.2 | 25.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.2 | 4.7 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 1.1 | 7.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.1 | 7.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.0 | 14.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 1.0 | 4.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.0 | 9.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 1.0 | 5.9 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 1.0 | 2.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.0 | 25.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 1.0 | 2.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.9 | 4.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.9 | 13.0 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.9 | 2.8 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.9 | 5.6 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.9 | 14.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.9 | 3.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.8 | 3.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.8 | 1.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.8 | 2.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.8 | 2.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.8 | 5.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.8 | 4.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.8 | 3.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.8 | 4.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.7 | 5.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.7 | 4.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.7 | 7.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.7 | 9.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.7 | 3.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.7 | 4.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.7 | 23.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.7 | 12.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.7 | 3.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.7 | 21.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 4.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 3.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.6 | 1.9 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.6 | 13.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 5.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.6 | 12.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 3.0 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.6 | 2.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.6 | 3.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.6 | 3.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.6 | 13.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.6 | 35.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.6 | 2.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.6 | 3.5 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.6 | 2.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.6 | 24.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.6 | 2.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 2.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 7.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.6 | 1.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.5 | 3.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.5 | 4.9 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.5 | 4.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 4.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 4.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 1.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.5 | 1.6 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.5 | 14.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 8.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 5.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.5 | 53.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.5 | 3.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.5 | 6.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 2.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 1.9 | GO:0051381 | histamine binding(GO:0051381) |
| 0.5 | 4.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 3.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 6.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.5 | 12.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 30.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.5 | 2.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.5 | 5.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 19.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 3.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.4 | 16.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.4 | 6.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 6.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 4.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 4.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 1.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 2.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 10.0 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.4 | 1.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 7.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 1.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 1.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.4 | 3.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 1.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 7.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.4 | 2.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 8.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 1.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 1.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 10.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 6.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 2.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.0 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.3 | 1.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 1.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 1.9 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.3 | 3.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.3 | 11.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 12.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 2.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 4.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.3 | 4.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 13.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 23.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.3 | 6.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 3.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 9.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 2.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 5.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.3 | 4.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 2.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 3.9 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 9.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 1.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.3 | 3.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 3.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 1.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 1.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 22.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 4.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 4.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 5.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 0.7 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.2 | 1.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 12.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.2 | 2.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 8.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 3.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 3.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 1.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 2.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.4 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 16.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.9 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 10.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 1.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 2.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 2.0 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.2 | 5.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 4.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 2.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 1.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.5 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.2 | 1.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.3 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.2 | 2.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 1.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 4.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 26.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 4.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 9.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 3.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 3.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 2.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.8 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 0.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 2.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 4.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 4.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 3.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 4.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 8.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 4.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 3.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.5 | GO:0004078 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 5.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.4 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.8 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 2.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 4.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.4 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.8 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 7.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 3.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 5.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 3.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 3.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 6.2 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 1.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 2.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 2.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 27.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 6.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 16.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 11.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 5.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 12.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 2.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 8.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 1.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 5.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 7.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 6.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 5.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 4.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 4.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 3.2 | GO:0042626 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.0 | 15.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.6 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 40.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.5 | 18.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.4 | 24.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 11.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 11.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 2.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 6.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 10.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 5.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 15.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 13.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 11.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 7.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 5.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 8.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 4.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 5.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 8.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 8.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 4.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 3.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 4.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 4.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 8.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 8.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 53.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 2.0 | 28.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.8 | 11.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 1.5 | 11.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 1.3 | 18.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.1 | 13.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.0 | 20.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.9 | 18.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.8 | 18.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 16.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.6 | 4.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 10.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 9.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.5 | 1.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 20.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.5 | 5.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.5 | 13.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.5 | 9.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.4 | 5.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 52.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.4 | 8.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 2.8 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.4 | 9.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 22.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 11.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 16.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 12.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.3 | 23.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 3.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 3.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 4.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 1.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 2.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 6.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 7.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 23.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 4.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 2.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 2.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 3.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 4.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 8.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 6.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 4.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 13.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 6.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 5.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.5 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 2.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 4.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 6.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 12.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 5.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 3.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 2.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 3.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |