Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
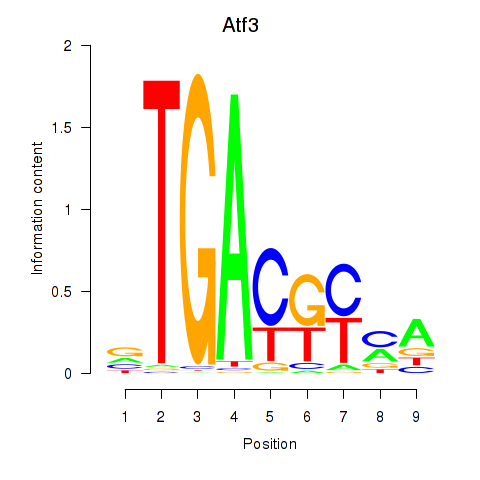
Results for Atf3
Z-value: 1.44
Transcription factors associated with Atf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf3
|
ENSMUSG00000026628.14 | Atf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf3 | mm39_v1_chr1_-_190915441_190915537 | -0.52 | 2.5e-06 | Click! |
Activity profile of Atf3 motif
Sorted Z-values of Atf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_94858417 | 7.12 |
ENSMUST00000038928.7
|
H1f9
|
H1.9 linker histone |
| chr9_-_106438798 | 6.27 |
ENSMUST00010126732.2
ENSMUST00010126033.2 ENSMUST00010181659.1 ENSMUST00010126065.2 ENSMUST00010126032.3 ENSMUST00000062917.16 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr4_+_39450265 | 5.91 |
ENSMUST00000029955.5
|
1700009N14Rik
|
RIKEN cDNA 1700009N14 gene |
| chr7_+_44145987 | 5.85 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr5_+_88127290 | 5.53 |
ENSMUST00000008051.5
|
Cabs1
|
calcium binding protein, spermatid specific 1 |
| chr8_+_55053809 | 5.51 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4 |
| chr6_+_88199250 | 5.09 |
ENSMUST00000061866.6
|
Dnajb8
|
DnaJ heat shock protein family (Hsp40) member B8 |
| chr7_+_44146029 | 4.97 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr17_-_56819422 | 4.95 |
ENSMUST00000052211.4
|
Znrf4
|
zinc and ring finger 4 |
| chr7_+_44146012 | 4.63 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr7_-_103778992 | 4.45 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr11_-_3672188 | 4.43 |
ENSMUST00000102950.10
ENSMUST00000101632.4 |
Osbp2
|
oxysterol binding protein 2 |
| chr14_+_70077841 | 4.28 |
ENSMUST00000022678.5
|
Pebp4
|
phosphatidylethanolamine binding protein 4 |
| chr6_+_41039255 | 4.28 |
ENSMUST00000103266.3
|
Trbv5
|
T cell receptor beta, variable 5 |
| chr4_+_104623505 | 4.25 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr14_+_64341735 | 4.11 |
ENSMUST00000022537.6
|
Prss52
|
protease, serine 52 |
| chr10_+_119828821 | 4.04 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr1_+_133291302 | 3.97 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr7_-_133203838 | 3.91 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr2_-_126625343 | 3.84 |
ENSMUST00000028842.9
ENSMUST00000130356.3 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr18_-_50701793 | 3.84 |
ENSMUST00000056460.4
|
Pudp
|
pseudouridine 5'-phosphatase |
| chr9_+_43978290 | 3.82 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr1_+_52047368 | 3.74 |
ENSMUST00000027277.7
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr4_-_63540653 | 3.73 |
ENSMUST00000102861.8
ENSMUST00000102862.4 |
Tex48
|
testis expressed 48 |
| chr7_-_133966588 | 3.69 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr9_-_106448182 | 3.67 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr2_+_80122811 | 3.64 |
ENSMUST00000057072.6
|
Prdx6b
|
peroxiredoxin 6B |
| chr9_+_43978369 | 3.62 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr16_-_16962279 | 3.62 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr6_+_49372463 | 3.62 |
ENSMUST00000024171.14
ENSMUST00000163954.8 ENSMUST00000172459.8 |
Stk31
|
serine threonine kinase 31 |
| chr4_+_56743407 | 3.60 |
ENSMUST00000095079.6
|
Actl7a
|
actin-like 7a |
| chr1_-_172085977 | 3.60 |
ENSMUST00000111243.2
|
Atp1a4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr13_-_23945189 | 3.55 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr9_+_27308087 | 3.55 |
ENSMUST00000214158.2
ENSMUST00000034473.7 |
Spata19
|
spermatogenesis associated 19 |
| chr6_+_113508636 | 3.51 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr18_-_3280999 | 3.50 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr4_-_34050038 | 3.50 |
ENSMUST00000084734.11
|
Spaca1
|
sperm acrosome associated 1 |
| chr19_-_11291805 | 3.47 |
ENSMUST00000187467.2
|
Ms4a14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr4_-_34050076 | 3.44 |
ENSMUST00000029927.6
|
Spaca1
|
sperm acrosome associated 1 |
| chr11_-_32477551 | 3.44 |
ENSMUST00000054327.3
|
Efcab9
|
EF-hand calcium binding domain 9 |
| chr11_+_115331365 | 3.43 |
ENSMUST00000093914.5
|
Trim80
|
tripartite motif-containing 80 |
| chr18_+_12874390 | 3.42 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr2_+_18703797 | 3.42 |
ENSMUST00000095132.10
|
Spag6
|
sperm associated antigen 6 |
| chr7_-_140367737 | 3.42 |
ENSMUST00000211616.2
ENSMUST00000026553.6 |
Syce1
|
synaptonemal complex central element protein 1 |
| chr6_+_41661356 | 3.40 |
ENSMUST00000031900.6
|
Llcfc1
|
LLLL and CFNLAS motif containing 1 |
| chr2_+_116709179 | 3.39 |
ENSMUST00000028834.3
|
Tmco5
|
transmembrane and coiled-coil domains 5 |
| chr3_+_130904000 | 3.38 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr17_-_41104954 | 3.33 |
ENSMUST00000131699.8
ENSMUST00000024724.14 ENSMUST00000232709.2 ENSMUST00000144243.8 |
Crisp2
|
cysteine-rich secretory protein 2 |
| chr9_+_106391771 | 3.30 |
ENSMUST00000085113.5
|
Iqcf5
|
IQ motif containing F5 |
| chr6_+_41523664 | 3.29 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr8_-_15096046 | 3.28 |
ENSMUST00000050493.4
ENSMUST00000123331.2 |
BB014433
|
expressed sequence BB014433 |
| chr11_-_77079794 | 3.28 |
ENSMUST00000108400.8
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chr9_-_71075939 | 3.26 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr4_+_42438970 | 3.21 |
ENSMUST00000238328.2
|
Gm21586
|
predicted gene, 21586 |
| chr4_+_42735545 | 3.21 |
ENSMUST00000068158.10
|
4930578G10Rik
|
RIKEN cDNA 4930578G10 gene |
| chr2_+_116709167 | 3.20 |
ENSMUST00000123598.8
ENSMUST00000155470.8 |
Tmco5
|
transmembrane and coiled-coil domains 5 |
| chr10_+_119828867 | 3.17 |
ENSMUST00000154238.8
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr7_-_133943703 | 3.16 |
ENSMUST00000106129.9
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr11_+_108286114 | 3.14 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr4_-_152533265 | 3.12 |
ENSMUST00000159840.8
ENSMUST00000105648.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr3_-_107462378 | 3.11 |
ENSMUST00000052853.8
|
Ubl4b
|
ubiquitin-like 4B |
| chr1_-_160986880 | 3.11 |
ENSMUST00000135643.8
ENSMUST00000178511.3 |
Tex50
|
testis expressed 50 |
| chr3_-_124374723 | 3.08 |
ENSMUST00000180162.8
ENSMUST00000047110.14 ENSMUST00000178485.8 |
1700003H04Rik
|
RIKEN cDNA 1700003H04 gene |
| chr11_-_103109247 | 3.07 |
ENSMUST00000103076.2
|
Spata32
|
spermatogenesis associated 32 |
| chr2_+_148631276 | 3.07 |
ENSMUST00000168443.8
ENSMUST00000028932.4 |
Cst12
|
cystatin 12 |
| chr11_+_87017878 | 3.06 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chr3_-_115923098 | 3.06 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr5_+_107655487 | 3.05 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr7_+_81823546 | 3.01 |
ENSMUST00000179318.8
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr6_+_32027216 | 2.99 |
ENSMUST00000031778.6
|
1700012A03Rik
|
RIKEN cDNA 1700012A03 gene |
| chr3_-_40856935 | 2.98 |
ENSMUST00000099123.5
|
1700034I23Rik
|
RIKEN cDNA 1700034I23 gene |
| chr17_-_47732798 | 2.98 |
ENSMUST00000073143.7
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chr4_-_41774097 | 2.94 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr8_+_22459785 | 2.92 |
ENSMUST00000070649.2
|
Ccdc70
|
coiled-coil domain containing 70 |
| chr12_+_113453131 | 2.92 |
ENSMUST00000063317.4
|
Adam6b
|
a disintegrin and metallopeptidase domain 6B |
| chr17_-_32639936 | 2.91 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr4_+_137989526 | 2.90 |
ENSMUST00000030539.10
|
Kif17
|
kinesin family member 17 |
| chr11_+_87018079 | 2.90 |
ENSMUST00000139532.2
|
Trim37
|
tripartite motif-containing 37 |
| chr7_-_103799746 | 2.88 |
ENSMUST00000059121.5
|
Ubqlnl
|
ubiquilin-like |
| chr13_+_65180860 | 2.86 |
ENSMUST00000099427.3
|
Spata31d1c
|
spermatogenesis associated 31 subfamily D, member 1C |
| chr6_+_103674695 | 2.85 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr16_-_10606513 | 2.85 |
ENSMUST00000051297.9
|
Tnp2
|
transition protein 2 |
| chr11_+_82782938 | 2.85 |
ENSMUST00000018988.6
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr1_-_186875270 | 2.83 |
ENSMUST00000184543.2
|
Spata17
|
spermatogenesis associated 17 |
| chr16_-_16962256 | 2.83 |
ENSMUST00000115711.10
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr19_+_41017714 | 2.83 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr15_-_78280099 | 2.82 |
ENSMUST00000229878.2
ENSMUST00000165170.8 ENSMUST00000074380.14 |
Tex33
|
testis expressed 33 |
| chr7_+_44926925 | 2.82 |
ENSMUST00000210861.2
|
Slc6a21
|
solute carrier family 6 member 21 |
| chr5_+_37332834 | 2.81 |
ENSMUST00000208827.2
ENSMUST00000207619.2 |
Gm1043
|
predicted gene 1043 |
| chr18_+_12874368 | 2.80 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr11_-_91468339 | 2.80 |
ENSMUST00000061019.6
|
Kif2b
|
kinesin family member 2B |
| chr11_-_52173391 | 2.79 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr2_-_3423646 | 2.79 |
ENSMUST00000064685.14
|
Meig1
|
meiosis expressed gene 1 |
| chr10_+_57521930 | 2.78 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr14_+_52155874 | 2.77 |
ENSMUST00000008957.13
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chrX_+_6934447 | 2.75 |
ENSMUST00000115751.3
|
Akap4
|
A kinase (PRKA) anchor protein 4 |
| chr3_-_94343874 | 2.74 |
ENSMUST00000204913.3
ENSMUST00000191506.8 ENSMUST00000199678.4 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr14_+_70815250 | 2.73 |
ENSMUST00000228554.2
|
Nudt18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr10_-_99595498 | 2.72 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr6_-_40906665 | 2.71 |
ENSMUST00000136499.2
|
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr19_-_58781709 | 2.71 |
ENSMUST00000166692.2
|
1700019N19Rik
|
RIKEN cDNA 1700019N19 gene |
| chr8_+_19244335 | 2.70 |
ENSMUST00000052601.4
|
Defb14
|
defensin beta 14 |
| chr16_-_50411484 | 2.68 |
ENSMUST00000062439.6
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr4_+_42091207 | 2.67 |
ENSMUST00000178882.2
|
Gm3893
|
predicted gene 3893 |
| chr4_+_140815644 | 2.66 |
ENSMUST00000051907.3
|
Spata21
|
spermatogenesis associated 21 |
| chr19_-_33369655 | 2.66 |
ENSMUST00000163093.2
|
Rnls
|
renalase, FAD-dependent amine oxidase |
| chr19_-_11261177 | 2.64 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr7_+_50248918 | 2.64 |
ENSMUST00000119710.3
|
4933405O20Rik
|
RIKEN cDNA 4933405O20 gene |
| chr7_-_24569130 | 2.62 |
ENSMUST00000122995.2
|
Lypd4
|
Ly6/Plaur domain containing 4 |
| chr19_-_6835538 | 2.60 |
ENSMUST00000113440.2
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr4_+_42293921 | 2.58 |
ENSMUST00000238203.2
|
Gm21953
|
predicted gene, 21953 |
| chr6_+_41112064 | 2.57 |
ENSMUST00000103272.4
|
Trbv14
|
T cell receptor beta, variable 14 |
| chr2_+_18703863 | 2.57 |
ENSMUST00000173763.2
|
Spag6
|
sperm associated antigen 6 |
| chr6_+_41095752 | 2.57 |
ENSMUST00000103269.3
|
Trbv12-2
|
T cell receptor beta, variable 12-2 |
| chr8_-_26087552 | 2.55 |
ENSMUST00000210234.2
ENSMUST00000211422.2 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr11_-_69508706 | 2.54 |
ENSMUST00000005334.3
|
Shbg
|
sex hormone binding globulin |
| chr8_+_117648474 | 2.54 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr8_-_70939964 | 2.54 |
ENSMUST00000045286.9
|
Tmem59l
|
transmembrane protein 59-like |
| chr2_+_59314989 | 2.54 |
ENSMUST00000028369.6
|
Dapl1
|
death associated protein-like 1 |
| chr5_-_123620632 | 2.53 |
ENSMUST00000198901.2
|
Il31
|
interleukin 31 |
| chr12_+_113507528 | 2.52 |
ENSMUST00000053086.3
|
Adam6a
|
a disintegrin and metallopeptidase domain 6A |
| chr8_-_26087475 | 2.52 |
ENSMUST00000210810.2
ENSMUST00000210616.2 ENSMUST00000079160.8 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr5_-_82271183 | 2.52 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr7_-_27146024 | 2.52 |
ENSMUST00000011895.14
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr7_+_127172701 | 2.50 |
ENSMUST00000121004.3
|
Phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr2_+_144665576 | 2.50 |
ENSMUST00000028918.4
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr16_+_17712061 | 2.46 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr8_+_26339646 | 2.45 |
ENSMUST00000098858.11
|
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr16_-_10213303 | 2.45 |
ENSMUST00000115831.2
|
Tekt5
|
tektin 5 |
| chr2_+_130248398 | 2.43 |
ENSMUST00000055421.6
|
Tmem239
|
transmembrane 239 |
| chr10_+_97528915 | 2.42 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr12_-_76293459 | 2.42 |
ENSMUST00000219327.2
ENSMUST00000021453.6 ENSMUST00000218426.2 |
Tex21
|
testis expressed gene 21 |
| chr12_-_28632514 | 2.41 |
ENSMUST00000110917.2
ENSMUST00000020965.14 |
Allc
|
allantoicase |
| chr4_+_43983472 | 2.41 |
ENSMUST00000095107.3
|
Ccin
|
calicin |
| chr5_-_103803578 | 2.40 |
ENSMUST00000120688.6
ENSMUST00000134926.8 ENSMUST00000120108.8 |
1700016H13Rik
|
RIKEN cDNA 1700016H13 gene |
| chr7_+_45271229 | 2.39 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr18_-_3281089 | 2.39 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr13_-_38220899 | 2.38 |
ENSMUST00000110233.8
ENSMUST00000074969.11 ENSMUST00000131066.2 |
Cage1
|
cancer antigen 1 |
| chr9_-_14411778 | 2.37 |
ENSMUST00000058796.7
|
Kdm4d
|
lysine (K)-specific demethylase 4D |
| chr12_-_85335193 | 2.36 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr4_-_128154709 | 2.36 |
ENSMUST00000053830.5
|
Hmgb4
|
high-mobility group box 4 |
| chr6_-_135287372 | 2.34 |
ENSMUST00000050471.4
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr11_+_105885461 | 2.34 |
ENSMUST00000190995.2
|
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr6_+_41098273 | 2.34 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr8_+_27937128 | 2.33 |
ENSMUST00000209669.2
|
Gm45861
|
predicted gene 45861 |
| chr8_-_73197616 | 2.32 |
ENSMUST00000019876.12
|
Calr3
|
calreticulin 3 |
| chr2_-_58990967 | 2.31 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chr7_+_67925718 | 2.29 |
ENSMUST00000210558.2
|
Fam169b
|
family with sequence similarity 169, member B |
| chr6_+_65929546 | 2.29 |
ENSMUST00000043382.9
|
4930544G11Rik
|
RIKEN cDNA 4930544G11 gene |
| chr17_+_27248233 | 2.29 |
ENSMUST00000053683.7
ENSMUST00000236222.2 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr3_+_134918298 | 2.29 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr17_+_48400153 | 2.29 |
ENSMUST00000233043.2
|
1700067P10Rik
|
RIKEN cDNA 1700067P10 gene |
| chr16_-_10213341 | 2.28 |
ENSMUST00000043415.13
|
Tekt5
|
tektin 5 |
| chr11_+_69231274 | 2.28 |
ENSMUST00000129321.2
|
Rnf227
|
ring finger protein 227 |
| chr2_-_25507680 | 2.28 |
ENSMUST00000028309.4
|
Ccdc183
|
coiled-coil domain containing 183 |
| chr2_+_25178147 | 2.27 |
ENSMUST00000028340.4
|
Tmem210
|
transmembrane protein 210 |
| chr8_-_13612397 | 2.26 |
ENSMUST00000187391.7
ENSMUST00000134023.9 ENSMUST00000151400.10 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr5_-_122270014 | 2.26 |
ENSMUST00000128101.8
ENSMUST00000132701.2 |
Ccdc63
|
coiled-coil domain containing 63 |
| chr9_-_35469818 | 2.25 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chr11_+_73220553 | 2.25 |
ENSMUST00000092926.11
ENSMUST00000117445.2 |
Spata22
Gm49340
|
spermatogenesis associated 22 predicted gene, 49340 |
| chrX_+_149330371 | 2.23 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr19_-_5553804 | 2.23 |
ENSMUST00000189704.2
|
Nscme3l
|
NSE3 homolog, SMC5-SMC6 complex component like |
| chr6_+_41025217 | 2.23 |
ENSMUST00000103264.3
|
Trbv3
|
T cell receptor beta, variable 3 |
| chr6_+_41092928 | 2.22 |
ENSMUST00000194399.2
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
| chr12_+_10419967 | 2.22 |
ENSMUST00000143739.9
ENSMUST00000002456.10 ENSMUST00000219826.2 ENSMUST00000217944.2 ENSMUST00000218339.3 ENSMUST00000118657.8 ENSMUST00000223534.2 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr15_-_36283244 | 2.22 |
ENSMUST00000228358.2
ENSMUST00000022890.10 |
Rnf19a
|
ring finger protein 19A |
| chr12_+_4132567 | 2.20 |
ENSMUST00000020986.15
ENSMUST00000049584.6 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr6_+_121277186 | 2.20 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr6_+_139987275 | 2.20 |
ENSMUST00000043797.6
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr3_-_121076745 | 2.19 |
ENSMUST00000135818.8
ENSMUST00000137234.2 |
Tlcd4
|
TLC domain containing 4 |
| chr8_+_96260694 | 2.19 |
ENSMUST00000041569.5
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr19_+_6135013 | 2.18 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr7_+_43090206 | 2.18 |
ENSMUST00000040227.3
|
Cldnd2
|
claudin domain containing 2 |
| chr6_-_129303659 | 2.18 |
ENSMUST00000203159.2
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr16_-_10608766 | 2.18 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr6_-_139987135 | 2.18 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1 |
| chr11_-_69214593 | 2.17 |
ENSMUST00000092973.6
|
Cntrob
|
centrobin, centrosomal BRCA2 interacting protein |
| chr17_+_46564457 | 2.17 |
ENSMUST00000233692.2
|
Lrrc73
|
leucine rich repeat containing 73 |
| chr6_-_23655130 | 2.16 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr11_+_31915542 | 2.16 |
ENSMUST00000102829.4
|
4930524B15Rik
|
RIKEN cDNA 4930524B15 gene |
| chr11_-_103395423 | 2.15 |
ENSMUST00000153273.3
|
Lrrc37a
|
leucine rich repeat containing 37A |
| chr2_-_110136074 | 2.15 |
ENSMUST00000046233.9
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr17_+_7592045 | 2.14 |
ENSMUST00000095726.11
ENSMUST00000128533.8 ENSMUST00000129709.8 ENSMUST00000147803.8 ENSMUST00000140192.8 ENSMUST00000138222.8 ENSMUST00000144861.2 |
Tcp10a
|
t-complex protein 10a |
| chr6_+_28427788 | 2.14 |
ENSMUST00000031719.7
|
Fscn3
|
fascin actin-bundling protein 3 |
| chr8_-_40964704 | 2.14 |
ENSMUST00000135269.8
ENSMUST00000034012.10 |
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr7_-_126548671 | 2.13 |
ENSMUST00000106339.2
ENSMUST00000052937.12 |
Asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr5_-_143211632 | 2.13 |
ENSMUST00000085733.9
|
Spdye4a
|
speedy/RINGO cell cycle regulator family, member E4A |
| chr14_-_21764638 | 2.13 |
ENSMUST00000073870.7
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr15_-_99864936 | 2.13 |
ENSMUST00000100209.6
|
Fam186a
|
family with sequence similarity 186, member A |
| chr9_+_106377181 | 2.11 |
ENSMUST00000085114.8
|
Iqcf1
|
IQ motif containing F1 |
| chr5_+_30486375 | 2.10 |
ENSMUST00000101448.5
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr7_+_44890497 | 2.10 |
ENSMUST00000213347.2
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr11_-_23469181 | 2.09 |
ENSMUST00000239488.2
ENSMUST00000020527.13 |
1700093K21Rik
|
RIKEN cDNA 1700093K21 gene |
| chr11_+_70506716 | 2.08 |
ENSMUST00000144960.2
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr17_+_34636321 | 2.07 |
ENSMUST00000142317.8
|
BC051142
|
cDNA sequence BC051142 |
| chr11_+_5578738 | 2.07 |
ENSMUST00000137933.2
|
Ankrd36
|
ankyrin repeat domain 36 |
| chr5_+_20112704 | 2.07 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_+_138794577 | 2.07 |
ENSMUST00000135509.2
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr5_+_105563605 | 2.06 |
ENSMUST00000112707.3
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr9_-_89996712 | 2.06 |
ENSMUST00000191353.2
ENSMUST00000085248.12 |
Morf4l1
|
mortality factor 4 like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.9 | 4.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.9 | 7.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.8 | 3.4 | GO:0061153 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.8 | 5.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.8 | 3.3 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.8 | 3.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.8 | 0.8 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.8 | 2.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.7 | 2.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.7 | 2.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.7 | 4.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.7 | 2.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) |
| 0.7 | 2.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.7 | 2.7 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.6 | 1.9 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.6 | 3.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.6 | 6.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.6 | 2.4 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.6 | 1.8 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.6 | 1.8 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.6 | 1.8 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.6 | 1.8 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.6 | 2.9 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.6 | 1.2 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.6 | 1.7 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.6 | 4.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.6 | 3.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.6 | 2.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.5 | 1.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.5 | 2.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 5.0 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.5 | 2.9 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.5 | 1.5 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.5 | 2.9 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.5 | 0.5 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.5 | 1.4 | GO:1902022 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine transport(GO:1902022) L-lysine import into cell(GO:1903410) |
| 0.5 | 5.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 7.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.5 | 1.9 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.5 | 0.5 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 0.5 | 6.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.5 | 0.5 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.5 | 5.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.5 | 5.5 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.5 | 1.4 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.5 | 2.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.5 | 1.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.4 | 4.9 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.4 | 2.2 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 | 1.7 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 1.3 | GO:0097037 | heme export(GO:0097037) |
| 0.4 | 3.4 | GO:0051324 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.4 | 1.7 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 4.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.4 | 1.6 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 1.2 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.4 | 1.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 2.4 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.4 | 3.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 0.8 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.4 | 2.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.4 | 1.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.4 | 3.0 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.4 | 2.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.4 | 2.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 1.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.4 | 2.9 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.4 | 6.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.4 | 1.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.4 | 1.8 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 | 2.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.4 | 8.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.4 | 0.7 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.4 | 0.4 | GO:1904023 | regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.3 | 1.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.3 | 2.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 1.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.3 | 1.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.3 | 6.6 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.3 | 1.0 | GO:0006571 | tyrosine biosynthetic process(GO:0006571) aromatic amino acid family biosynthetic process(GO:0009073) aromatic amino acid family biosynthetic process, prephenate pathway(GO:0009095) |
| 0.3 | 3.7 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 3.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 2.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.3 | 1.0 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.3 | 1.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 2.5 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.3 | 1.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 0.6 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.3 | 0.6 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.3 | 2.2 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.3 | 0.3 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.3 | 1.2 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.3 | 1.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 0.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.3 | 1.2 | GO:0045575 | basophil activation(GO:0045575) |
| 0.3 | 1.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.3 | 1.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 0.9 | GO:2000554 | T-helper 1 cell cytokine production(GO:0035744) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.3 | 2.4 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.3 | 0.9 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.3 | 5.0 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.3 | 1.5 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.3 | 0.6 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 3.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 10.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 4.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 3.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 1.4 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.3 | 1.9 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.3 | 0.8 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.3 | 0.8 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 | 1.1 | GO:0061187 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 8.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.3 | 1.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 1.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.3 | 0.5 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.3 | 2.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.3 | 1.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.3 | 0.8 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.3 | 0.5 | GO:0090649 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 1.6 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.3 | 1.3 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.3 | 0.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.3 | 1.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 2.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 2.5 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 1.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.2 | 1.4 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 | 0.5 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.2 | 1.2 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.2 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.2 | 0.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 1.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 0.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 2.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 1.1 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.2 | 1.8 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.2 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.2 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 1.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.9 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 1.8 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 5.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.2 | 2.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 1.8 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 0.7 | GO:0006235 | dTDP biosynthetic process(GO:0006233) dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTDP metabolic process(GO:0046072) dTTP metabolic process(GO:0046075) |
| 0.2 | 0.4 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 2.8 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 29.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 0.9 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.2 | 0.6 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 | 5.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 0.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 5.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 0.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 4.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.8 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.2 | 2.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.6 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 2.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 1.4 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.2 | 1.4 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.2 | 0.4 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 1.8 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 0.6 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 | 2.3 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.2 | 0.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.2 | 2.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 2.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 5.1 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.2 | 2.3 | GO:0090220 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.2 | 0.6 | GO:0061723 | glycophagy(GO:0061723) |
| 0.2 | 1.3 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 0.4 | GO:0072708 | response to sorbitol(GO:0072708) |
| 0.2 | 7.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 1.3 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.2 | 0.5 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.2 | 1.8 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 | 0.2 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.2 | 1.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.5 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.2 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 0.5 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 0.9 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 0.7 | GO:0015755 | fructose transport(GO:0015755) |
| 0.2 | 3.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.8 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.8 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.2 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.2 | 0.5 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.7 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.2 | 0.6 | GO:0032329 | serine transport(GO:0032329) |
| 0.2 | 2.4 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 0.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 0.5 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.2 | 3.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 3.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 3.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 1.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.6 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 1.7 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.2 | 1.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 1.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 2.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 1.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.5 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 2.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.6 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 1.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.0 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.4 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 1.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.4 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 1.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.6 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.9 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.5 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 1.0 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.6 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.9 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.6 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.1 | 0.9 | GO:0072603 | interleukin-5 secretion(GO:0072603) interleukin-13 secretion(GO:0072611) regulation of interleukin-5 secretion(GO:2000662) positive regulation of interleukin-5 secretion(GO:2000664) regulation of interleukin-13 secretion(GO:2000665) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 2.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.4 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 1.3 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.1 | 0.9 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.8 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 7.2 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.5 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 1.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 1.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.5 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 0.9 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 1.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.2 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.1 | 0.5 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.3 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 1.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.6 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.3 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.3 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.5 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 1.4 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 44.0 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.4 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 1.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.7 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 0.4 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 3.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.5 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.5 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.3 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.1 | 1.3 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 1.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.6 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 1.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.5 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.4 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.4 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 0.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.1 | 1.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 2.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 2.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 1.9 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.6 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 2.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.6 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 1.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 3.0 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 1.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.5 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 1.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.5 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.7 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 4.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.9 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.1 | 0.5 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 1.7 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 0.4 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.7 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 2.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.2 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 0.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 1.0 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.0 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 0.5 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.1 | 1.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.2 | GO:1901738 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 1.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.6 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.1 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.1 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.9 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.3 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.1 | 1.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.2 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.1 | 0.3 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.3 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 2.8 | GO:0098840 | protein transport along microtubule(GO:0098840) |
| 0.1 | 0.6 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.5 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.9 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.6 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.5 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.7 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.1 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 2.1 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 1.1 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 3.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.3 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.0 | 1.5 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 2.5 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 2.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.4 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.8 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 1.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 3.9 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 1.7 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 1.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.4 | GO:0072710 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 1.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.6 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 1.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.8 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.9 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 1.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 2.1 | GO:0006304 | DNA modification(GO:0006304) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.7 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.5 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.8 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.0 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.6 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.4 | GO:0090023 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 1.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.4 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 2.7 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.4 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 4.8 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.3 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 3.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.8 | 4.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.7 | 3.9 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.6 | 1.9 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.6 | 5.6 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.6 | 3.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.5 | 4.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 6.1 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 4.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 2.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.4 | 1.3 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.4 | 1.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 1.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 1.6 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.4 | 12.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 1.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 1.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.4 | 2.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 1.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.3 | 2.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.3 | 2.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 14.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 6.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 7.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 5.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 0.9 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 3.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 3.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 3.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 1.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.3 | 0.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 1.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.3 | 1.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 1.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 2.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 1.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 1.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 2.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 4.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 2.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 6.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 3.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.0 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 0.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 1.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 1.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 1.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.6 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 0.9 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 0.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.9 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 0.5 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.2 | 1.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 7.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 1.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 5.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 1.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 0.6 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 0.8 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 2.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 5.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.4 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 0.9 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 1.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 3.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 19.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.7 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 1.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 6.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.5 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.5 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 7.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 8.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 1.5 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 2.0 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.1 | 3.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 6.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 0.8 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 2.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 2.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 6.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 6.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.1 | 2.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.2 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 0.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.9 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 47.2 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 2.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 2.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 2.3 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 0.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.5 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 3.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.6 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 3.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 1.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 1.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.4 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 1.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 1.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 1.1 | 3.4 | GO:0050354 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 1.0 | 2.9 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.9 | 4.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.9 | 2.7 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.8 | 2.3 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.7 | 2.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.7 | 5.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 2.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.6 | 2.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.6 | 3.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.6 | 2.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.6 | 2.3 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 0.6 | 2.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 3.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 2.9 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.5 | 1.9 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.4 | 3.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 1.3 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.4 | 2.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 1.6 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.4 | 2.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.4 | 2.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 3.9 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.4 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 2.7 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.5 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.4 | 2.2 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.4 | 2.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.4 | 1.8 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.4 | 1.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.4 | 2.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 1.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 1.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 1.0 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.3 | 3.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 10.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.3 | 5.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 2.0 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 0.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.3 | 1.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 2.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 0.9 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.3 | 1.6 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 0.9 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 0.6 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.3 | 1.5 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.3 | 0.9 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 1.6 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.3 | 1.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 1.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.3 | 0.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.3 | 0.8 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.3 | 0.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.3 | 8.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 3.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.2 | 1.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 2.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 1.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.2 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) |
| 0.2 | 1.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 0.5 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 1.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 0.7 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) uniporter activity(GO:0015292) |
| 0.2 | 0.5 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 3.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 1.8 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.2 | 0.7 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.2 | 2.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 3.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.9 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 0.4 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.2 | 0.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 4.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.6 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.2 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 1.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 0.6 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 2.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 1.6 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.2 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 0.6 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 2.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 4.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 1.1 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.2 | 5.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.7 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 1.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 4.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 1.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 6.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.2 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 2.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.7 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 2.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 0.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 0.5 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.2 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 1.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 0.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 0.5 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 1.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 18.0 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 2.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 4.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.7 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.4 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 4.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 6.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 3.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.3 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 0.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 2.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.6 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 9.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 6.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.7 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 1.8 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 0.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.0 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 4.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 5.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 7.0 | GO:0032934 | sterol binding(GO:0032934) |
| 0.1 | 1.3 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 1.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.5 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 2.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.2 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 2.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 2.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.4 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.7 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.2 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.5 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.9 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 3.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 4.8 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 1.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.2 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.4 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 1.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.2 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 0.2 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 3.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.2 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 2.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.2 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.1 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 9.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 1.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 3.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 4.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.6 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 1.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.9 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 1.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 4.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) U4atac snRNA binding(GO:0030622) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.8 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 3.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.6 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 4.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 3.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.4 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 6.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.0 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 8.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 9.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 5.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 4.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 6.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.5 | 1.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 4.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 5.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 2.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 2.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 5.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 7.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 2.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 2.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 7.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 5.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 4.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 3.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 4.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 5.5 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 1.8 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 1.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 4.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |