Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Atf4
Z-value: 0.99
Transcription factors associated with Atf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf4
|
ENSMUSG00000042406.9 | Atf4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf4 | mm39_v1_chr15_+_80139371_80139386 | 0.01 | 9.3e-01 | Click! |
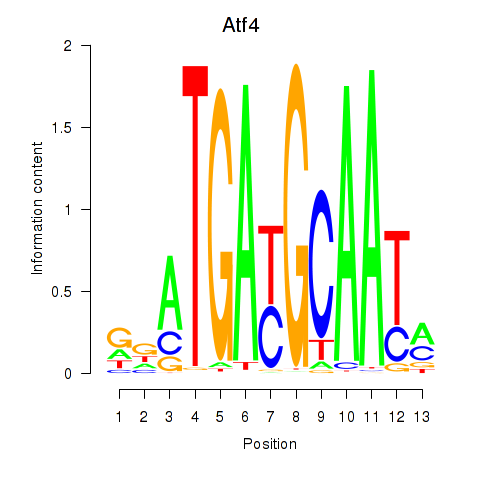
Activity profile of Atf4 motif
Sorted Z-values of Atf4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_119181703 | 7.79 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr8_+_86219191 | 5.91 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr15_+_9335636 | 5.16 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr11_-_75313350 | 5.14 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_-_75313412 | 4.78 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr19_-_43512929 | 4.28 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr7_+_44117511 | 3.75 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr10_-_127358231 | 3.48 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_+_44117444 | 3.47 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr10_-_127358300 | 3.42 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr11_+_96162283 | 3.31 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chr7_+_44117475 | 3.28 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chrX_+_102465616 | 3.05 |
ENSMUST00000182089.2
|
Gm26992
|
predicted gene, 26992 |
| chr7_+_44117404 | 2.96 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr9_+_21746785 | 2.68 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr7_+_51528715 | 2.59 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr9_+_32283779 | 2.57 |
ENSMUST00000047334.10
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr11_+_67061908 | 2.52 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr13_-_53627110 | 2.50 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr10_-_83369247 | 2.44 |
ENSMUST00000146640.8
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr8_+_95113066 | 2.41 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr11_+_67061837 | 2.40 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr11_-_120534469 | 2.31 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr14_-_32407203 | 2.23 |
ENSMUST00000096038.4
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene |
| chr1_-_180021039 | 2.09 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr7_+_51528788 | 1.97 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr13_-_30729242 | 1.86 |
ENSMUST00000042834.4
|
Uqcrfs1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr4_+_129083553 | 1.74 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr11_-_53321606 | 1.71 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr6_+_129510117 | 1.69 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr18_-_64649497 | 1.59 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr18_-_64649620 | 1.59 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr7_-_143153785 | 1.56 |
ENSMUST00000105909.4
ENSMUST00000010899.14 |
Cars
|
cysteinyl-tRNA synthetase |
| chr11_+_9141934 | 1.46 |
ENSMUST00000042740.13
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr8_+_111760521 | 1.44 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr9_-_44255456 | 1.42 |
ENSMUST00000077353.15
|
Hmbs
|
hydroxymethylbilane synthase |
| chr6_+_129510145 | 1.42 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr14_+_55777807 | 1.37 |
ENSMUST00000226352.2
|
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr1_+_185095232 | 1.36 |
ENSMUST00000046514.13
|
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr11_-_101062111 | 1.36 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr7_-_3551003 | 1.35 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr11_-_115167775 | 1.31 |
ENSMUST00000021078.3
|
Fdxr
|
ferredoxin reductase |
| chr3_-_89905927 | 1.29 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr15_+_99291100 | 1.29 |
ENSMUST00000159209.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr16_+_22737227 | 1.27 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr5_+_118307754 | 1.25 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr13_+_49835576 | 1.24 |
ENSMUST00000165316.8
ENSMUST00000047363.14 |
Iars
|
isoleucine-tRNA synthetase |
| chr3_-_127202693 | 1.23 |
ENSMUST00000182078.9
|
Ank2
|
ankyrin 2, brain |
| chr7_-_109092834 | 1.22 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr1_-_71692320 | 1.19 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr3_-_127202663 | 1.12 |
ENSMUST00000182008.8
ENSMUST00000182547.8 |
Ank2
|
ankyrin 2, brain |
| chr10_+_127126643 | 1.06 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr13_+_49835667 | 1.05 |
ENSMUST00000172254.3
|
Iars
|
isoleucine-tRNA synthetase |
| chr3_-_127202635 | 1.04 |
ENSMUST00000182959.8
|
Ank2
|
ankyrin 2, brain |
| chr13_-_101829070 | 1.01 |
ENSMUST00000187009.7
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr3_-_127202586 | 1.00 |
ENSMUST00000183095.3
ENSMUST00000182610.8 |
Ank2
|
ankyrin 2, brain |
| chr15_+_99291491 | 0.99 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr6_+_129510331 | 0.96 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr11_-_53321242 | 0.95 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr16_-_23751144 | 0.95 |
ENSMUST00000231038.2
|
Rtp2
|
receptor transporter protein 2 |
| chrX_+_163221035 | 0.91 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr14_-_36857083 | 0.89 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr11_-_58425662 | 0.88 |
ENSMUST00000213188.3
|
Olfr330
|
olfactory receptor 330 |
| chr14_-_50521663 | 0.88 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr2_+_3514071 | 0.88 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr9_+_109760856 | 0.88 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr5_-_90371816 | 0.86 |
ENSMUST00000118816.6
ENSMUST00000048363.10 |
Cox18
|
cytochrome c oxidase assembly protein 18 |
| chr5_-_148336574 | 0.84 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_-_20282684 | 0.82 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr1_+_156666485 | 0.82 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr12_+_108859557 | 0.81 |
ENSMUST00000221377.2
|
Wdr25
|
WD repeat domain 25 |
| chr3_-_59038539 | 0.81 |
ENSMUST00000198838.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr4_+_138606671 | 0.81 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr13_-_101829132 | 0.80 |
ENSMUST00000035532.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr6_+_55014973 | 0.80 |
ENSMUST00000003572.10
|
Gars
|
glycyl-tRNA synthetase |
| chr15_+_98367777 | 0.79 |
ENSMUST00000216180.2
|
OR5BS1P
|
olfactory receptor 240, pseudogene 1 |
| chr8_+_72795062 | 0.77 |
ENSMUST00000215324.2
|
Olfr372
|
olfactory receptor 372 |
| chr7_+_79910948 | 0.76 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr12_-_108859123 | 0.74 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr13_-_118523760 | 0.74 |
ENSMUST00000022245.10
|
Mrps30
|
mitochondrial ribosomal protein S30 |
| chr3_-_89905547 | 0.73 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr5_-_145077048 | 0.68 |
ENSMUST00000031627.9
|
Pdap1
|
PDGFA associated protein 1 |
| chr15_+_102012782 | 0.67 |
ENSMUST00000230474.2
|
Tns2
|
tensin 2 |
| chr14_-_56339915 | 0.67 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr7_+_109617456 | 0.67 |
ENSMUST00000084731.5
|
Ipo7
|
importin 7 |
| chr19_-_50667079 | 0.66 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr2_-_18053158 | 0.66 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chr10_+_100176698 | 0.63 |
ENSMUST00000168769.3
|
Gm4302
|
predicted gene 4302 |
| chr6_+_65019793 | 0.63 |
ENSMUST00000204696.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_+_103980178 | 0.63 |
ENSMUST00000170919.2
|
Gm21293
|
predicted gene, 21293 |
| chr10_+_104031258 | 0.63 |
ENSMUST00000163113.3
|
Gm4340
|
predicted gene 4340 |
| chr10_+_104014239 | 0.63 |
ENSMUST00000179258.2
|
Gm21312
|
predicted gene, 21312 |
| chr10_+_104022748 | 0.63 |
ENSMUST00000177792.2
|
Gm20765
|
predicted gene, 20765 |
| chr15_+_99590098 | 0.62 |
ENSMUST00000228185.2
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr10_+_100181834 | 0.61 |
ENSMUST00000218922.2
|
Gm4303
|
predicted gene 4303 |
| chr10_+_100186955 | 0.61 |
ENSMUST00000219159.2
|
Gm4305
|
predicted gene 4305 |
| chr10_+_100197200 | 0.61 |
ENSMUST00000177945.2
|
Gm4307
|
predicted gene 4307 |
| chr2_-_18053595 | 0.61 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr16_-_36154692 | 0.61 |
ENSMUST00000114850.3
|
Cstdc6
|
cystatin domain containing 6 |
| chr9_-_15023396 | 0.61 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1 |
| chr5_+_42225303 | 0.60 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr15_+_99291455 | 0.60 |
ENSMUST00000162624.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr5_-_148336711 | 0.59 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_-_87811788 | 0.59 |
ENSMUST00000216461.2
ENSMUST00000217112.2 |
Olfr464
|
olfactory receptor 464 |
| chr3_-_96170627 | 0.58 |
ENSMUST00000171473.3
|
H4c14
|
H4 clustered histone 14 |
| chr7_+_140043283 | 0.56 |
ENSMUST00000211093.3
ENSMUST00000216258.2 ENSMUST00000215308.2 ENSMUST00000217179.2 |
Olfr533
|
olfactory receptor 533 |
| chr8_+_106427774 | 0.55 |
ENSMUST00000098444.9
ENSMUST00000212352.2 |
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr19_-_40576817 | 0.54 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr10_+_121575819 | 0.53 |
ENSMUST00000065600.8
ENSMUST00000136432.2 |
BC048403
|
cDNA sequence BC048403 |
| chr7_-_139956244 | 0.50 |
ENSMUST00000216727.2
|
Olfr530
|
olfactory receptor 530 |
| chr4_+_117692583 | 0.44 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr5_+_93241385 | 0.43 |
ENSMUST00000201421.4
ENSMUST00000202415.4 ENSMUST00000202217.3 |
Septin11
|
septin 11 |
| chr11_-_87716849 | 0.42 |
ENSMUST00000103177.10
|
Lpo
|
lactoperoxidase |
| chr14_-_8068465 | 0.41 |
ENSMUST00000215479.2
|
Olfr721-ps1
|
olfactory receptor 721, pseudogene 1 |
| chr19_-_40576897 | 0.41 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr7_-_103543692 | 0.40 |
ENSMUST00000081748.6
|
Olfr64
|
olfactory receptor 64 |
| chr2_-_89423470 | 0.39 |
ENSMUST00000217254.2
ENSMUST00000217192.2 ENSMUST00000213221.2 |
Olfr1246
|
olfactory receptor 1246 |
| chr19_-_40576782 | 0.39 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chrX_+_93278203 | 0.39 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15 |
| chr11_+_100211363 | 0.39 |
ENSMUST00000152521.2
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr12_+_87762282 | 0.37 |
ENSMUST00000221442.2
|
Eif1ad13
|
eukaryotic translation initiation factor 1A domain containing 13 |
| chr7_+_144450260 | 0.36 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr2_+_87853118 | 0.36 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr14_-_54655237 | 0.35 |
ENSMUST00000195999.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr16_+_36097505 | 0.35 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr14_+_54422156 | 0.33 |
ENSMUST00000103707.2
|
Traj34
|
T cell receptor alpha joining 34 |
| chr18_+_40390013 | 0.33 |
ENSMUST00000096572.2
ENSMUST00000236889.2 |
Kctd16
|
potassium channel tetramerisation domain containing 16 |
| chr4_-_118036827 | 0.32 |
ENSMUST00000097911.9
|
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr1_-_78465479 | 0.32 |
ENSMUST00000190441.2
ENSMUST00000170217.8 ENSMUST00000188247.7 ENSMUST00000068333.14 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr14_-_50538979 | 0.25 |
ENSMUST00000216195.2
ENSMUST00000214372.2 ENSMUST00000214756.2 |
Olfr733
|
olfactory receptor 733 |
| chr10_-_68114543 | 0.25 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr19_-_8690330 | 0.24 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr6_-_97436223 | 0.24 |
ENSMUST00000113359.8
|
Frmd4b
|
FERM domain containing 4B |
| chr7_-_140087224 | 0.22 |
ENSMUST00000209873.2
ENSMUST00000064392.8 ENSMUST00000215768.2 ENSMUST00000215340.2 |
Olfr536
|
olfactory receptor 536 |
| chr1_-_31017049 | 0.18 |
ENSMUST00000178317.4
|
Gm5698
|
predicted gene 5698 |
| chr11_+_57899890 | 0.16 |
ENSMUST00000071487.13
ENSMUST00000178636.2 |
Larp1
|
La ribonucleoprotein domain family, member 1 |
| chr3_-_59038160 | 0.16 |
ENSMUST00000197841.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr5_+_93241287 | 0.13 |
ENSMUST00000074733.11
ENSMUST00000201700.4 ENSMUST00000202196.4 ENSMUST00000202308.4 |
Septin11
|
septin 11 |
| chr15_-_76191301 | 0.13 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr1_-_190915441 | 0.12 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr12_+_85646162 | 0.10 |
ENSMUST00000050687.14
|
Jdp2
|
Jun dimerization protein 2 |
| chr1_-_92412835 | 0.10 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr2_-_37007795 | 0.08 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr3_+_137849560 | 0.07 |
ENSMUST00000159622.8
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr12_+_73333553 | 0.04 |
ENSMUST00000140523.8
ENSMUST00000126488.8 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr9_-_79700660 | 0.03 |
ENSMUST00000034878.12
|
Tmem30a
|
transmembrane protein 30A |
| chr11_-_98291220 | 0.01 |
ENSMUST00000128897.2
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr7_+_138448308 | 0.00 |
ENSMUST00000155672.8
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr4_-_41774097 | 0.00 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 1.5 | 5.9 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 1.4 | 4.3 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.8 | 2.5 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.8 | 9.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.8 | 2.4 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.8 | 2.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.6 | 3.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.6 | 2.9 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.6 | 7.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.5 | 4.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 | 1.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.5 | 1.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.5 | 3.7 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.4 | 2.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.4 | 2.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.4 | 1.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.4 | 2.7 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.3 | 2.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 0.8 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.3 | 4.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 4.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.4 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 1.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.2 | 0.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 0.8 | GO:0070127 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 5.2 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.1 | 0.4 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 3.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 4.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 2.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 13.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.7 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.9 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.4 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.4 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.2 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.8 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 1.0 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.4 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 1.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 4.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 3.3 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 3.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.6 | 4.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.6 | 2.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.6 | 9.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 1.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 4.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.4 | 1.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 4.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 4.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 4.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 7.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 3.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 1.4 | 4.3 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.3 | 7.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.2 | 5.9 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.8 | 2.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.8 | 3.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.8 | 2.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.6 | 4.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 2.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.5 | 1.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.5 | 1.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.5 | 2.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 1.4 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.4 | 1.3 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.3 | 2.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.8 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 4.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 0.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 2.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 5.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 5.1 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 13.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.8 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 9.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 4.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 1.5 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 4.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 12.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 9.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 3.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 4.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |