Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
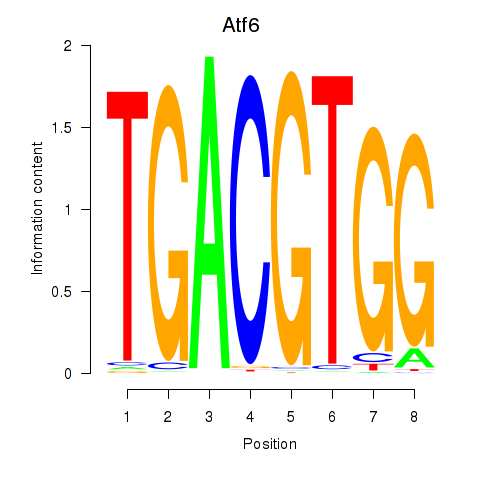
Results for Atf6
Z-value: 1.20
Transcription factors associated with Atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf6
|
ENSMUSG00000026663.7 | Atf6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf6 | mm39_v1_chr1_-_170695328_170695354 | 0.39 | 6.3e-04 | Click! |
Activity profile of Atf6 motif
Sorted Z-values of Atf6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_126721878 | 14.21 |
ENSMUST00000046765.10
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr8_+_126722113 | 13.67 |
ENSMUST00000212831.2
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr8_-_65146079 | 10.95 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr5_+_32293145 | 8.07 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr16_+_94171477 | 6.87 |
ENSMUST00000117648.9
ENSMUST00000147352.8 ENSMUST00000150346.8 ENSMUST00000155692.8 ENSMUST00000153988.9 ENSMUST00000139513.9 ENSMUST00000141856.8 ENSMUST00000152117.8 ENSMUST00000150097.8 ENSMUST00000122895.8 ENSMUST00000151770.8 ENSMUST00000231569.2 ENSMUST00000147046.8 ENSMUST00000149885.8 ENSMUST00000127667.8 ENSMUST00000119131.3 ENSMUST00000145883.2 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chrX_+_135567124 | 6.87 |
ENSMUST00000060904.11
ENSMUST00000113100.2 ENSMUST00000128040.2 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chrX_+_40490005 | 6.37 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chrX_+_80114242 | 6.29 |
ENSMUST00000171953.8
ENSMUST00000026760.3 |
Tmem47
|
transmembrane protein 47 |
| chr10_+_29087658 | 6.13 |
ENSMUST00000213489.2
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr6_+_7555053 | 5.95 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr6_-_126717114 | 5.59 |
ENSMUST00000112242.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chrX_+_151922936 | 5.55 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr10_+_29087602 | 5.50 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr1_-_175319842 | 5.47 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr14_-_55150547 | 5.40 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr13_+_58955506 | 5.32 |
ENSMUST00000079828.7
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr18_-_31580436 | 5.31 |
ENSMUST00000025110.5
|
Syt4
|
synaptotagmin IV |
| chr10_-_18619658 | 5.26 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr17_-_37334240 | 5.10 |
ENSMUST00000102665.11
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr13_+_58956077 | 5.07 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr16_+_41353360 | 5.06 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr18_+_23937019 | 5.06 |
ENSMUST00000025127.5
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr13_-_105191403 | 4.97 |
ENSMUST00000063551.7
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein |
| chr7_+_126549692 | 4.88 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr17_-_37334091 | 4.85 |
ENSMUST00000167275.3
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr10_-_49659355 | 4.82 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr5_-_118382926 | 4.81 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr4_+_85123654 | 4.80 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr6_-_126717590 | 4.62 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr5_+_137059127 | 4.53 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr15_-_81845019 | 4.51 |
ENSMUST00000230229.2
|
Pmm1
|
phosphomannomutase 1 |
| chr6_-_124441731 | 4.51 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr16_-_44153288 | 4.49 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr5_+_110692162 | 4.46 |
ENSMUST00000040001.14
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr15_-_81845050 | 4.46 |
ENSMUST00000071462.7
ENSMUST00000023112.12 |
Pmm1
|
phosphomannomutase 1 |
| chr7_+_126549859 | 4.39 |
ENSMUST00000106333.8
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr8_-_70939964 | 4.36 |
ENSMUST00000045286.9
|
Tmem59l
|
transmembrane protein 59-like |
| chr5_-_52628825 | 4.36 |
ENSMUST00000198008.5
ENSMUST00000059428.7 |
Ccdc149
|
coiled-coil domain containing 149 |
| chr19_+_16933471 | 4.36 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr2_+_121125918 | 4.35 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr16_-_44153498 | 4.33 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr11_-_75686874 | 4.23 |
ENSMUST00000021209.8
|
Doc2b
|
double C2, beta |
| chr15_-_102419115 | 4.19 |
ENSMUST00000171565.8
|
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr8_+_114932312 | 4.17 |
ENSMUST00000049509.7
ENSMUST00000150963.2 |
Vat1l
|
vesicle amine transport protein 1 like |
| chr6_+_77219627 | 4.11 |
ENSMUST00000159616.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr7_+_126550009 | 4.03 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chrX_-_133709733 | 4.03 |
ENSMUST00000035559.11
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr6_+_77219698 | 4.00 |
ENSMUST00000161677.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr4_-_138820269 | 3.99 |
ENSMUST00000042844.7
|
Nbl1
|
NBL1, DAN family BMP antagonist |
| chr4_+_85123358 | 3.96 |
ENSMUST00000107188.10
|
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr16_+_17577493 | 3.94 |
ENSMUST00000165790.9
|
Klhl22
|
kelch-like 22 |
| chr16_+_17577464 | 3.91 |
ENSMUST00000129199.8
|
Klhl22
|
kelch-like 22 |
| chr7_+_97437709 | 3.83 |
ENSMUST00000206984.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr13_+_58955675 | 3.75 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chrX_+_165127688 | 3.69 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr11_-_74480870 | 3.65 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr6_-_136150076 | 3.48 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr1_-_55265925 | 3.43 |
ENSMUST00000027121.15
ENSMUST00000114428.3 |
Rftn2
|
raftlin family member 2 |
| chr3_-_127019496 | 3.37 |
ENSMUST00000182064.9
ENSMUST00000182452.8 |
Ank2
|
ankyrin 2, brain |
| chr7_-_125681577 | 3.35 |
ENSMUST00000073935.7
|
Gsg1l
|
GSG1-like |
| chr6_-_136150491 | 3.30 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr5_+_137286535 | 3.29 |
ENSMUST00000024099.11
ENSMUST00000196208.5 ENSMUST00000085934.4 |
Ache
|
acetylcholinesterase |
| chr1_+_115612458 | 3.27 |
ENSMUST00000043725.9
|
Cntnap5a
|
contactin associated protein-like 5A |
| chr2_+_67578556 | 3.25 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr16_+_41353212 | 3.25 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr7_-_45391879 | 3.23 |
ENSMUST00000210754.2
ENSMUST00000210147.2 |
Sult2b1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr8_+_13034245 | 3.14 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr19_-_5040344 | 3.14 |
ENSMUST00000056129.9
|
Npas4
|
neuronal PAS domain protein 4 |
| chr8_+_26210484 | 3.06 |
ENSMUST00000210629.2
|
Plpp5
|
phospholipid phosphatase 5 |
| chr5_-_51725059 | 3.06 |
ENSMUST00000127135.3
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr4_+_127066667 | 3.00 |
ENSMUST00000106094.9
|
Dlgap3
|
DLG associated protein 3 |
| chr12_-_4891435 | 2.98 |
ENSMUST00000219880.2
ENSMUST00000020964.7 |
Fkbp1b
|
FK506 binding protein 1b |
| chr2_+_129854256 | 2.96 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr13_+_93440265 | 2.95 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_50966670 | 2.91 |
ENSMUST00000081851.4
|
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chrX_+_133618693 | 2.91 |
ENSMUST00000113201.8
ENSMUST00000051256.10 ENSMUST00000113199.8 ENSMUST00000035748.14 ENSMUST00000113198.8 ENSMUST00000113197.2 |
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chrX_-_135642025 | 2.88 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr13_-_92268156 | 2.88 |
ENSMUST00000151408.9
ENSMUST00000216219.2 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr6_-_8778439 | 2.85 |
ENSMUST00000115520.8
ENSMUST00000038403.12 ENSMUST00000115518.8 |
Ica1
|
islet cell autoantigen 1 |
| chrX_-_135641869 | 2.83 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr4_-_110209106 | 2.76 |
ENSMUST00000106603.9
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chrX_+_100342749 | 2.69 |
ENSMUST00000118111.8
ENSMUST00000130555.8 ENSMUST00000151528.8 |
Nlgn3
|
neuroligin 3 |
| chr12_+_82663660 | 2.65 |
ENSMUST00000161801.8
ENSMUST00000185665.7 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr2_+_84564394 | 2.63 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr10_-_18619439 | 2.60 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr12_-_36092475 | 2.55 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr18_+_63110899 | 2.54 |
ENSMUST00000025474.14
|
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr5_+_137059522 | 2.51 |
ENSMUST00000187382.2
|
Vgf
|
VGF nerve growth factor inducible |
| chr9_+_122980006 | 2.50 |
ENSMUST00000026890.6
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr2_+_102380357 | 2.44 |
ENSMUST00000028612.8
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr2_-_105229653 | 2.43 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr1_+_143516402 | 2.40 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr8_+_26210064 | 2.35 |
ENSMUST00000068916.16
ENSMUST00000139836.8 |
Plpp5
|
phospholipid phosphatase 5 |
| chr3_+_131270648 | 2.35 |
ENSMUST00000199878.5
ENSMUST00000196408.5 ENSMUST00000200527.5 |
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr3_+_131270529 | 2.34 |
ENSMUST00000029666.14
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr6_-_83513222 | 2.33 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr12_+_82663347 | 2.33 |
ENSMUST00000186848.5
|
Rgs6
|
regulator of G-protein signaling 6 |
| chr11_-_20781009 | 2.32 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr6_-_8778106 | 2.31 |
ENSMUST00000151758.2
ENSMUST00000115519.8 ENSMUST00000153390.8 |
Ica1
|
islet cell autoantigen 1 |
| chr7_+_45434755 | 2.30 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr12_+_82663785 | 2.29 |
ENSMUST00000200911.4
|
Rgs6
|
regulator of G-protein signaling 6 |
| chr18_+_63110924 | 2.25 |
ENSMUST00000150267.2
ENSMUST00000236925.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr18_+_63111005 | 2.25 |
ENSMUST00000235372.2
ENSMUST00000237483.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chrX_-_72830487 | 2.20 |
ENSMUST00000052761.9
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD+), gamma |
| chr4_+_138181616 | 2.17 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr12_-_72117865 | 2.13 |
ENSMUST00000050649.6
|
Gpr135
|
G protein-coupled receptor 135 |
| chr3_+_103482591 | 2.13 |
ENSMUST00000090697.11
ENSMUST00000239027.2 |
Syt6
|
synaptotagmin VI |
| chr14_-_60324265 | 2.11 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr3_-_126955976 | 2.10 |
ENSMUST00000182994.8
|
Ank2
|
ankyrin 2, brain |
| chr6_-_118456198 | 2.10 |
ENSMUST00000161170.2
|
Zfp9
|
zinc finger protein 9 |
| chr5_+_30310138 | 2.07 |
ENSMUST00000058045.5
|
Garem2
|
GRB2 associated regulator of MAPK1 subtype 2 |
| chr6_-_83513184 | 2.07 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr9_-_95632387 | 2.07 |
ENSMUST00000189137.7
ENSMUST00000053785.10 |
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr4_+_125384481 | 2.04 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr17_+_46608842 | 2.03 |
ENSMUST00000166617.8
ENSMUST00000170271.2 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr7_+_110376859 | 1.98 |
ENSMUST00000148292.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr2_-_79287095 | 1.96 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr7_-_105289515 | 1.96 |
ENSMUST00000133519.8
ENSMUST00000209550.2 ENSMUST00000210911.2 ENSMUST00000084782.10 ENSMUST00000131446.8 |
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr9_+_121589044 | 1.95 |
ENSMUST00000093772.4
|
Zfp651
|
zinc finger protein 651 |
| chr6_-_115228800 | 1.95 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr11_-_6425877 | 1.94 |
ENSMUST00000179343.3
|
Purb
|
purine rich element binding protein B |
| chr11_+_92990110 | 1.93 |
ENSMUST00000107863.4
|
Car10
|
carbonic anhydrase 10 |
| chr15_+_82159094 | 1.93 |
ENSMUST00000116423.3
ENSMUST00000230418.2 |
Septin3
|
septin 3 |
| chr1_-_125839897 | 1.92 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr11_-_70578905 | 1.92 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr16_+_20514925 | 1.91 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr17_-_24424456 | 1.88 |
ENSMUST00000201583.2
ENSMUST00000202925.4 ENSMUST00000167791.9 ENSMUST00000201960.4 ENSMUST00000040474.11 ENSMUST00000201089.4 ENSMUST00000201301.4 ENSMUST00000201805.4 ENSMUST00000168410.9 ENSMUST00000097376.10 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chrX_+_100342813 | 1.85 |
ENSMUST00000065858.3
|
Nlgn3
|
neuroligin 3 |
| chr3_+_123061094 | 1.84 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr8_-_68270936 | 1.84 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_+_29433247 | 1.83 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr7_+_15832383 | 1.83 |
ENSMUST00000006181.7
|
Napa
|
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
| chr7_-_105282687 | 1.82 |
ENSMUST00000147044.4
ENSMUST00000106791.8 ENSMUST00000153371.9 ENSMUST00000106789.8 |
Trim3
|
tripartite motif-containing 3 |
| chr18_-_43032535 | 1.79 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr18_-_61344644 | 1.78 |
ENSMUST00000146409.8
|
Slc26a2
|
solute carrier family 26 (sulfate transporter), member 2 |
| chrX_+_20736405 | 1.77 |
ENSMUST00000115342.10
ENSMUST00000009530.5 |
Timp1
|
tissue inhibitor of metalloproteinase 1 |
| chr14_+_3575406 | 1.75 |
ENSMUST00000124353.2
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr5_-_116729870 | 1.74 |
ENSMUST00000076124.7
|
Srrm4
|
serine/arginine repetitive matrix 4 |
| chr17_+_87415049 | 1.72 |
ENSMUST00000041369.8
ENSMUST00000234803.2 |
Socs5
|
suppressor of cytokine signaling 5 |
| chr11_-_70578775 | 1.69 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chrX_-_7242065 | 1.67 |
ENSMUST00000191497.2
ENSMUST00000115744.2 |
Usp27x
|
ubiquitin specific peptidase 27, X chromosome |
| chrX_+_72830607 | 1.67 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr8_-_70929555 | 1.67 |
ENSMUST00000066597.13
ENSMUST00000210250.2 ENSMUST00000209415.2 ENSMUST00000166976.3 |
Klhl26
|
kelch-like 26 |
| chr11_-_29975916 | 1.66 |
ENSMUST00000058902.6
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr6_+_71470987 | 1.66 |
ENSMUST00000114179.3
|
Rnf103
|
ring finger protein 103 |
| chr15_+_79400597 | 1.65 |
ENSMUST00000010974.9
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr16_+_20551853 | 1.65 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr4_+_43406435 | 1.65 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr4_+_46489248 | 1.64 |
ENSMUST00000030018.5
|
Nans
|
N-acetylneuraminic acid synthase (sialic acid synthase) |
| chr5_-_136199525 | 1.60 |
ENSMUST00000041048.6
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr19_+_45006552 | 1.59 |
ENSMUST00000237043.2
ENSMUST00000178087.3 |
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chrX_-_20816841 | 1.59 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr13_+_118851214 | 1.57 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chrX_+_72830668 | 1.56 |
ENSMUST00000002090.3
|
Ssr4
|
signal sequence receptor, delta |
| chr6_-_82751429 | 1.56 |
ENSMUST00000000642.11
|
Hk2
|
hexokinase 2 |
| chr3_+_134534754 | 1.55 |
ENSMUST00000029822.6
|
Tacr3
|
tachykinin receptor 3 |
| chr4_+_8535604 | 1.53 |
ENSMUST00000060232.8
|
Rab2a
|
RAB2A, member RAS oncogene family |
| chr4_-_118401185 | 1.51 |
ENSMUST00000128098.8
|
Tmem125
|
transmembrane protein 125 |
| chr18_+_84738996 | 1.50 |
ENSMUST00000235504.2
|
Dipk1c
|
divergent protein kinase domain 1C |
| chr11_+_87938128 | 1.49 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr5_+_142946098 | 1.49 |
ENSMUST00000031565.15
ENSMUST00000198017.5 |
Fscn1
|
fascin actin-bundling protein 1 |
| chr11_-_97590460 | 1.48 |
ENSMUST00000103148.8
ENSMUST00000169807.8 |
Pcgf2
|
polycomb group ring finger 2 |
| chr14_+_4230658 | 1.46 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr6_-_101354858 | 1.45 |
ENSMUST00000075994.11
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr18_-_34505544 | 1.45 |
ENSMUST00000236887.2
|
Reep5
|
receptor accessory protein 5 |
| chr15_+_82159398 | 1.45 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chr2_+_80145805 | 1.44 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr15_+_80861966 | 1.43 |
ENSMUST00000139517.9
ENSMUST00000137255.3 ENSMUST00000137004.2 |
Sgsm3
|
small G protein signaling modulator 3 |
| chr14_-_124914516 | 1.42 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr10_+_128158413 | 1.40 |
ENSMUST00000219836.2
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr18_-_43032514 | 1.39 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr7_-_105282751 | 1.38 |
ENSMUST00000057525.14
|
Trim3
|
tripartite motif-containing 3 |
| chr4_-_47474283 | 1.38 |
ENSMUST00000044148.3
|
Alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chrX_-_133501874 | 1.37 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr18_+_34994253 | 1.37 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr1_-_161078723 | 1.36 |
ENSMUST00000051925.5
ENSMUST00000071718.12 |
Prdx6
|
peroxiredoxin 6 |
| chr18_+_69654572 | 1.36 |
ENSMUST00000200862.4
|
Tcf4
|
transcription factor 4 |
| chr5_+_4073343 | 1.36 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr13_+_93440572 | 1.34 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_133109059 | 1.34 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr17_-_34250474 | 1.32 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr10_+_128158328 | 1.32 |
ENSMUST00000219037.2
ENSMUST00000026446.4 |
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr9_+_119274009 | 1.31 |
ENSMUST00000035094.14
|
Exog
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr2_+_156563284 | 1.29 |
ENSMUST00000099145.6
|
Dlgap4
|
DLG associated protein 4 |
| chr17_-_34250616 | 1.28 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr4_+_132262853 | 1.27 |
ENSMUST00000094657.10
ENSMUST00000105940.10 ENSMUST00000105939.10 ENSMUST00000150207.8 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr6_+_17693941 | 1.26 |
ENSMUST00000115420.8
ENSMUST00000115419.8 |
St7
|
suppression of tumorigenicity 7 |
| chr19_+_23118545 | 1.26 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9 |
| chr19_+_10872587 | 1.23 |
ENSMUST00000025642.14
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr18_+_69654231 | 1.23 |
ENSMUST00000202350.4
ENSMUST00000202477.4 |
Tcf4
|
transcription factor 4 |
| chr5_-_136199482 | 1.23 |
ENSMUST00000196454.5
ENSMUST00000197052.2 |
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr6_+_17694003 | 1.23 |
ENSMUST00000052113.12
ENSMUST00000081635.13 |
St7
|
suppression of tumorigenicity 7 |
| chr7_-_44519196 | 1.23 |
ENSMUST00000046575.17
|
Ptov1
|
prostate tumor over expressed gene 1 |
| chr3_+_122688721 | 1.21 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr4_-_57300361 | 1.20 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr5_+_31855394 | 1.20 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr16_-_57051727 | 1.19 |
ENSMUST00000226586.2
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chr7_-_45116316 | 1.19 |
ENSMUST00000033093.10
|
Bax
|
BCL2-associated X protein |
| chr19_-_58444336 | 1.18 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr18_+_34464185 | 1.17 |
ENSMUST00000072576.10
ENSMUST00000119329.8 |
Srp19
|
signal recognition particle 19 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 26.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 3.6 | 10.9 | GO:0030070 | insulin processing(GO:0030070) |
| 2.4 | 14.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 2.2 | 9.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.8 | 5.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.5 | 4.5 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 1.5 | 6.0 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 1.4 | 6.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 1.2 | 4.7 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.1 | 12.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.1 | 3.3 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 1.1 | 3.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.0 | 3.1 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 1.0 | 3.1 | GO:1904633 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 1.0 | 4.0 | GO:0090027 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of monocyte chemotaxis(GO:0090027) |
| 1.0 | 9.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.0 | 2.9 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 1.0 | 3.8 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.7 | 2.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.7 | 1.4 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.6 | 5.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.6 | 5.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.6 | 5.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 1.6 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.5 | 5.5 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.5 | 2.9 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.5 | 1.9 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.5 | 3.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.5 | 1.4 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.4 | 4.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 2.9 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.4 | 1.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 4.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 8.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.4 | 8.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.4 | 7.0 | GO:0043084 | penile erection(GO:0043084) |
| 0.4 | 2.7 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 3.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.3 | 1.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.3 | 1.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 | 1.6 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.3 | 1.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.3 | 7.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 2.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 3.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.9 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.3 | 13.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 10.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 0.9 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.3 | 2.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 4.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 | 3.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 1.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.3 | 7.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 2.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 1.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 0.7 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 2.1 | GO:0061092 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 0.6 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 0.6 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.2 | 4.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 2.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.2 | 1.0 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.2 | 1.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.2 | 1.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 1.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.7 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 0.6 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 2.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.5 | GO:1902871 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 1.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 1.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 1.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 1.5 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 1.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 0.5 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 1.2 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 1.8 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 6.1 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.1 | 3.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 6.2 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 1.3 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.1 | 0.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 1.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 5.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 3.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 2.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 2.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 4.3 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 5.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 2.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.4 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 2.0 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 4.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.4 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.1 | 1.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.1 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.1 | 0.4 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 4.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.7 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 5.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.6 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 3.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.3 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.5 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 6.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.6 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 3.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.8 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 1.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 1.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.7 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.0 | 1.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.3 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.4 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 1.0 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 1.1 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.8 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 4.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 1.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.7 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 3.8 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 9.4 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.5 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 1.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 27.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.1 | 5.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.0 | 3.1 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 1.0 | 7.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.0 | 2.9 | GO:0097144 | BAX complex(GO:0097144) |
| 0.9 | 16.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.8 | 16.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 1.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.6 | 4.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 1.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.4 | 1.7 | GO:0071920 | cleavage body(GO:0071920) |
| 0.3 | 2.6 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 1.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 1.5 | GO:0044393 | microspike(GO:0044393) |
| 0.3 | 4.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 0.3 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.2 | 3.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 14.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 5.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 11.8 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 10.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 1.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 0.9 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 0.6 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 1.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 3.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 0.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 4.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 12.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 2.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 8.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.0 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 3.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 5.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 10.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.7 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.5 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 7.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 19.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 2.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 3.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 4.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 8.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 8.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 19.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 4.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 4.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 3.6 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 3.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 2.4 | 14.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 1.8 | 8.9 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.8 | 5.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.4 | 5.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.3 | 4.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.3 | 26.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.2 | 4.7 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.1 | 6.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.1 | 6.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.1 | 3.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.1 | 6.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.9 | 10.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.9 | 6.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.7 | 2.2 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.5 | 3.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) steroid sulfotransferase activity(GO:0050294) |
| 0.5 | 1.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.4 | 2.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 2.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 1.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 2.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 2.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 1.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 1.7 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.3 | 4.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 10.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 4.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 1.0 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.3 | 0.9 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.3 | 0.8 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.3 | 3.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 6.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 4.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 1.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 1.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 5.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 5.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 9.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 7.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 2.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.4 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 1.6 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.6 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 1.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 3.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 1.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 2.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 2.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 1.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 4.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.4 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 3.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 1.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 3.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 7.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 3.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.8 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 1.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 8.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 9.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 1.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.9 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 5.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 2.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 3.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 3.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 3.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.8 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 1.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 5.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 14.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 6.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 3.7 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.5 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.6 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 7.4 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 13.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 12.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 9.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 6.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 10.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 6.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 2.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 3.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 8.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 26.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.6 | 7.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.5 | 10.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.5 | 8.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 11.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 14.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.4 | 2.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.4 | 4.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 5.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 10.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 8.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 4.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 4.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 12.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 10.6 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 2.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 2.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |