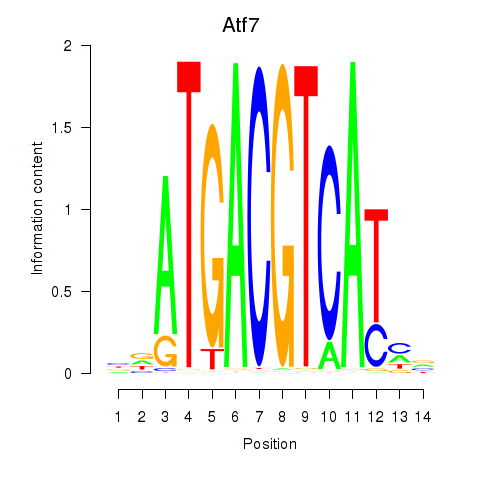
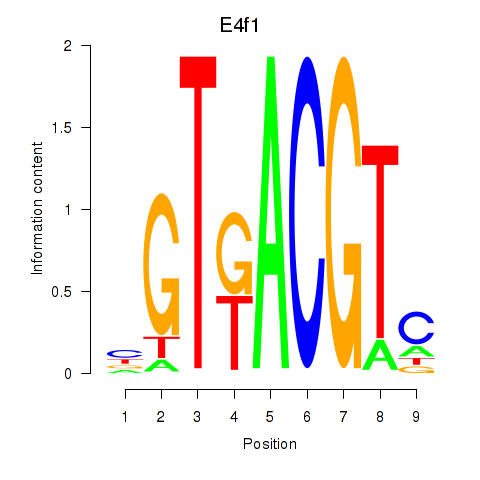
Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Atf7_E4f1
Z-value: 0.93
Transcription factors associated with Atf7_E4f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf7
|
ENSMUSG00000099083.8 | Atf7 |
|
Atf7
|
ENSMUSG00000099083.8 | Atf7 |
|
E4f1
|
ENSMUSG00000024137.10 | E4f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf7 | mm39_v1_chr15_-_102533870_102533936 | 0.63 | 3.7e-09 | Click! |
| E4f1 | mm39_v1_chr17_-_24674252_24674366 | 0.62 | 8.6e-09 | Click! |
Activity profile of Atf7_E4f1 motif
Sorted Z-values of Atf7_E4f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf7_E4f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 1.3 | 2.7 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 1.0 | 3.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.0 | 5.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.8 | 3.4 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.8 | 3.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.8 | 4.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.8 | 7.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.8 | 3.0 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.8 | 3.8 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.7 | 14.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.7 | 3.9 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.7 | 2.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.6 | 3.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.6 | 1.7 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.5 | 1.6 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.5 | 2.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.5 | 6.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.5 | 2.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.5 | 1.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.5 | 3.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.5 | 3.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.5 | 0.9 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 2.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.4 | 1.3 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.4 | 1.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.4 | 2.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.4 | 1.2 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.4 | 3.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.4 | 2.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 1.3 | GO:2000705 | positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 0.3 | 1.3 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.3 | 1.0 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.3 | 1.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.3 | 1.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 0.9 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.3 | 4.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.8 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.3 | 4.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 0.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 1.3 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 | 2.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 1.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.3 | 1.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 0.8 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.3 | 1.0 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.3 | 0.8 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.3 | 0.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.3 | 3.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.3 | 0.8 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 0.5 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 4.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.2 | 1.9 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 0.7 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 0.7 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.2 | 1.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 0.7 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.2 | 1.8 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 3.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 1.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 | 0.7 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.2 | 1.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 2.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.8 | GO:0006272 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.2 | 0.8 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 1.6 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 2.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 0.8 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 3.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 4.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.7 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.2 | 2.6 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 | 4.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 2.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 3.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 0.5 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.2 | 0.9 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 3.5 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.2 | 2.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 0.6 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.2 | 0.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.4 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 2.5 | GO:0030432 | peristalsis(GO:0030432) |
| 0.1 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 4.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 2.9 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.7 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.9 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 1.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 1.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 4.9 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.1 | 0.8 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 | 0.4 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.1 | 0.5 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 4.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 2.5 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 3.6 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.5 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 1.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.8 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.6 | GO:0097167 | circadian regulation of translation(GO:0097167) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 1.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.2 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 2.8 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.3 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.1 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 4.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.2 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 | 1.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 1.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 | 1.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.2 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.1 | 1.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.9 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 1.3 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 1.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.9 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 1.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.5 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.8 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.6 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.1 | 0.2 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 1.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.8 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 2.9 | GO:0090077 | foam cell differentiation(GO:0090077) |
| 0.1 | 0.6 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 1.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 2.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.3 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 0.5 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 1.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.8 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.1 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 0.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 6.9 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.1 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.0 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.7 | GO:0072401 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.1 | 0.6 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.8 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.6 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 1.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.7 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.2 | GO:1903011 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.4 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 1.9 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 5.1 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) positive regulation of cortisol secretion(GO:0051464) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 2.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.8 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.6 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.5 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 2.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 1.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.2 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 1.4 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.5 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 7.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 3.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 2.0 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.5 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.4 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 2.0 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 2.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.3 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.3 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 3.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:0044393 | microspike(GO:0044393) |
| 0.7 | 3.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 2.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 2.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.4 | 6.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.4 | 5.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 5.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.4 | 3.9 | GO:0070187 | telosome(GO:0070187) |
| 0.4 | 1.8 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.3 | 2.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 8.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 1.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 0.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 0.8 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.3 | 5.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 2.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 0.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 6.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 0.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 4.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 1.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 4.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 4.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 2.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 2.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 1.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 1.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 2.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 4.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 3.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 4.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 7.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.9 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 2.0 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.8 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 3.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 2.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.9 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 2.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 4.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 3.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 3.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.8 | GO:0048787 | early phagosome(GO:0032009) presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 4.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 2.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 5.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.6 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 5.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 6.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.1 | 3.4 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.9 | 3.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.8 | 6.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 2.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.7 | 2.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.6 | 2.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.6 | 1.8 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.6 | 2.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.6 | 3.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.6 | 6.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 3.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.5 | 1.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.5 | 1.8 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 2.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.4 | 3.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.4 | 3.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 1.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 1.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 0.9 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.3 | 0.9 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.3 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 1.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 0.8 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.3 | 2.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.0 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 1.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 1.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 3.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 0.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 0.4 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 0.6 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 3.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 0.7 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.8 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 1.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 3.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 2.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 2.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.3 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 2.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 4.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.3 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 10.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 1.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 3.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.8 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.5 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 2.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 5.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 3.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 5.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 1.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 3.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 2.8 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 3.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 4.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 5.8 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 4.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 3.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 3.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.9 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 4.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 2.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 3.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 8.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 5.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 10.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 9.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 9.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 3.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 14.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 3.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 5.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 4.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 3.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 6.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 11.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 6.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 3.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 2.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 4.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 2.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 3.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |