Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Atoh1_Bhlhe23
Z-value: 0.86
Transcription factors associated with Atoh1_Bhlhe23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
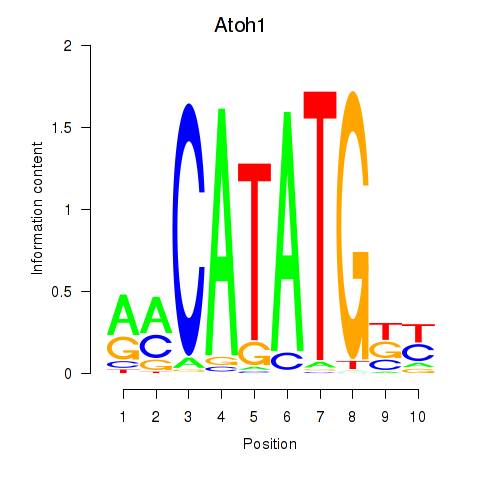
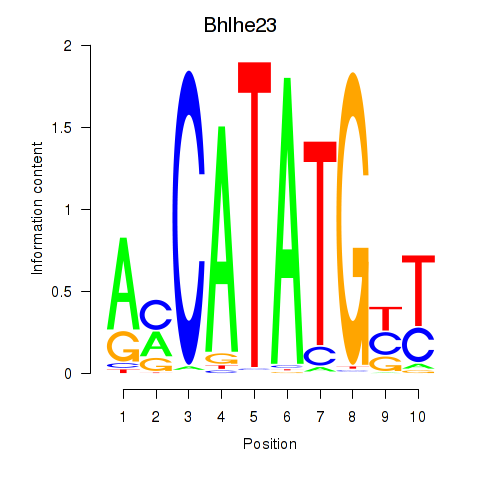
Atoh1
|
ENSMUSG00000073043.6 | Atoh1 |
|
Bhlhe23
|
ENSMUSG00000045493.5 | Bhlhe23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atoh1 | mm39_v1_chr6_+_64706101_64706117 | -0.32 | 6.5e-03 | Click! |
| Bhlhe23 | mm39_v1_chr2_-_180418693_180418693 | 0.17 | 1.4e-01 | Click! |
Activity profile of Atoh1_Bhlhe23 motif
Sorted Z-values of Atoh1_Bhlhe23 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atoh1_Bhlhe23
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_165127688 | 12.27 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr2_+_61876956 | 11.05 |
ENSMUST00000112480.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_61876923 | 10.84 |
ENSMUST00000054484.15
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr10_-_70491764 | 10.57 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr2_+_61876806 | 10.39 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_-_79959178 | 8.48 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_126792056 | 8.13 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr3_-_59127571 | 7.61 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr2_-_53975501 | 5.29 |
ENSMUST00000100089.3
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr16_-_31133622 | 5.21 |
ENSMUST00000115230.2
ENSMUST00000130560.8 |
Apod
|
apolipoprotein D |
| chr2_-_79959802 | 5.11 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_-_110149916 | 4.78 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr10_-_25076008 | 4.45 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr16_-_44153288 | 4.40 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr16_-_44153498 | 4.35 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr10_+_69542153 | 4.29 |
ENSMUST00000182992.8
|
Ank3
|
ankyrin 3, epithelial |
| chr5_-_124325213 | 4.20 |
ENSMUST00000161938.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr11_-_69493567 | 4.03 |
ENSMUST00000138694.2
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr1_+_177272215 | 4.02 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr15_-_98707367 | 3.87 |
ENSMUST00000230409.2
|
Ddn
|
dendrin |
| chr2_+_65760477 | 3.87 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr15_-_27788693 | 3.70 |
ENSMUST00000226287.2
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr11_-_75686874 | 3.68 |
ENSMUST00000021209.8
|
Doc2b
|
double C2, beta |
| chr15_+_30172716 | 3.61 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr5_-_142594549 | 3.47 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr6_+_17743581 | 3.46 |
ENSMUST00000000674.13
ENSMUST00000077080.9 |
St7
|
suppression of tumorigenicity 7 |
| chr2_-_25471703 | 3.43 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr1_-_131441962 | 3.41 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr7_-_126303689 | 3.30 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr7_-_126303887 | 3.21 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr11_+_58845502 | 3.06 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr9_-_99599312 | 3.06 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr7_+_127503812 | 3.00 |
ENSMUST00000151451.3
ENSMUST00000124533.3 ENSMUST00000206745.2 ENSMUST00000206140.2 |
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr2_+_173918715 | 2.99 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr19_-_37153436 | 2.99 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr5_+_63957805 | 2.88 |
ENSMUST00000162757.4
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr7_-_126303947 | 2.84 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr14_+_43951187 | 2.83 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr3_-_84167119 | 2.79 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chr2_+_21372338 | 2.72 |
ENSMUST00000055946.8
|
Gpr158
|
G protein-coupled receptor 158 |
| chr11_-_98220466 | 2.62 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr14_-_70945434 | 2.56 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr17_-_47143214 | 2.56 |
ENSMUST00000233537.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr1_+_177272297 | 2.47 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr7_-_104646109 | 2.39 |
ENSMUST00000050599.5
|
Olfr672
|
olfactory receptor 672 |
| chr2_-_174188505 | 2.33 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr6_-_113478779 | 2.30 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr2_-_71198091 | 2.27 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr7_+_30157704 | 2.24 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr15_-_89726063 | 2.24 |
ENSMUST00000029441.4
|
Syt10
|
synaptotagmin X |
| chr11_-_76468527 | 2.14 |
ENSMUST00000176179.8
|
Abr
|
active BCR-related gene |
| chr2_+_25293140 | 2.03 |
ENSMUST00000154809.8
ENSMUST00000055921.14 ENSMUST00000141567.8 |
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr11_-_101308441 | 1.92 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr5_-_52628825 | 1.87 |
ENSMUST00000198008.5
ENSMUST00000059428.7 |
Ccdc149
|
coiled-coil domain containing 149 |
| chr2_-_29142965 | 1.87 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr11_-_35871300 | 1.81 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr15_-_75963446 | 1.81 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chrX_+_55500170 | 1.81 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr2_+_59442378 | 1.71 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_-_121637505 | 1.70 |
ENSMUST00000138157.8
|
Frmd5
|
FERM domain containing 5 |
| chr13_+_37529184 | 1.65 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr18_+_65158873 | 1.61 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr8_-_55340024 | 1.59 |
ENSMUST00000176866.8
|
Wdr17
|
WD repeat domain 17 |
| chr6_-_12749409 | 1.51 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr2_+_81883566 | 1.47 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr9_+_24194729 | 1.44 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr6_-_12749192 | 1.43 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr1_+_9978863 | 1.22 |
ENSMUST00000052843.12
ENSMUST00000171802.8 ENSMUST00000125294.9 ENSMUST00000140948.9 |
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr18_+_34892599 | 1.22 |
ENSMUST00000097622.4
|
Fam53c
|
family with sequence similarity 53, member C |
| chr7_+_55443931 | 1.20 |
ENSMUST00000205796.2
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr11_-_115494692 | 1.19 |
ENSMUST00000125097.2
ENSMUST00000019135.14 ENSMUST00000106508.10 |
Gga3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr18_+_65022035 | 1.19 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr2_-_79287095 | 1.18 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr5_-_74229021 | 1.17 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr11_+_73051228 | 1.13 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr7_+_55443893 | 1.11 |
ENSMUST00000032627.5
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr1_-_150268470 | 1.10 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chrX_+_35592006 | 1.09 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr5_-_66775979 | 1.06 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr13_+_108182941 | 1.04 |
ENSMUST00000224361.2
ENSMUST00000224390.2 |
Smim15
|
small integral membrane protein 15 |
| chr5_-_24652775 | 1.03 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr11_+_49138278 | 1.03 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr11_+_78394273 | 1.02 |
ENSMUST00000001130.8
ENSMUST00000125670.3 |
Sebox
|
SEBOX homeobox |
| chr1_+_177273226 | 0.97 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr7_+_6463510 | 0.95 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chr7_+_127503251 | 0.92 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr1_-_170133901 | 0.91 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr10_+_61010983 | 0.91 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr2_-_156022054 | 0.86 |
ENSMUST00000126992.8
ENSMUST00000146288.8 ENSMUST00000029149.13 ENSMUST00000109587.9 ENSMUST00000109584.8 |
Rbm39
|
RNA binding motif protein 39 |
| chr15_+_101071948 | 0.86 |
ENSMUST00000000544.12
|
Acvr1b
|
activin A receptor, type 1B |
| chr6_+_112250719 | 0.85 |
ENSMUST00000032376.6
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chrX_+_158491589 | 0.85 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_-_87295292 | 0.83 |
ENSMUST00000067692.13
|
Rad51c
|
RAD51 paralog C |
| chr6_-_119925387 | 0.80 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr11_+_120123727 | 0.79 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr9_+_106158212 | 0.78 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A |
| chr9_+_26910995 | 0.78 |
ENSMUST00000213770.2
ENSMUST00000213683.2 ENSMUST00000039161.10 |
Thyn1
|
thymocyte nuclear protein 1 |
| chr11_+_32592707 | 0.76 |
ENSMUST00000109366.8
ENSMUST00000093205.13 ENSMUST00000076383.8 |
Fbxw11
|
F-box and WD-40 domain protein 11 |
| chr1_-_52839530 | 0.73 |
ENSMUST00000159725.2
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr15_+_84553801 | 0.69 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr5_+_75312939 | 0.69 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr16_+_95058895 | 0.64 |
ENSMUST00000113859.8
ENSMUST00000152516.2 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_-_90487583 | 0.64 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr17_-_31363245 | 0.63 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr16_-_17540685 | 0.62 |
ENSMUST00000232163.2
ENSMUST00000232202.2 ENSMUST00000080936.14 ENSMUST00000232645.2 ENSMUST00000232431.2 |
Med15
|
mediator complex subunit 15 |
| chr2_+_89804937 | 0.60 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr16_-_17540805 | 0.59 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr11_-_72441054 | 0.57 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr9_+_19716202 | 0.55 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr8_-_107792264 | 0.55 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr1_+_150268544 | 0.53 |
ENSMUST00000124973.9
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr9_+_19620729 | 0.53 |
ENSMUST00000217450.4
ENSMUST00000212013.4 |
Olfr857
|
olfactory receptor 857 |
| chr10_+_61011164 | 0.50 |
ENSMUST00000140456.2
|
Tbata
|
thymus, brain and testes associated |
| chr4_+_136150835 | 0.49 |
ENSMUST00000088677.5
ENSMUST00000121571.8 ENSMUST00000117699.2 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr9_-_71803354 | 0.49 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr13_-_99584091 | 0.47 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chr18_+_74349189 | 0.47 |
ENSMUST00000025444.8
|
Cxxc1
|
CXXC finger 1 (PHD domain) |
| chr11_+_68393845 | 0.47 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr7_+_28488380 | 0.43 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr1_+_156386414 | 0.42 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr2_+_127429125 | 0.39 |
ENSMUST00000028852.13
|
Mrps5
|
mitochondrial ribosomal protein S5 |
| chr17_-_26011241 | 0.37 |
ENSMUST00000237093.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr7_+_17799863 | 0.37 |
ENSMUST00000108487.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr7_+_107501834 | 0.33 |
ENSMUST00000210420.2
|
Olfr472
|
olfactory receptor 472 |
| chr7_+_17799889 | 0.30 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chrX_+_36059274 | 0.30 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr15_-_91075933 | 0.27 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr3_-_151468529 | 0.25 |
ENSMUST00000046739.6
|
Ifi44l
|
interferon-induced protein 44 like |
| chr17_-_26011357 | 0.24 |
ENSMUST00000236683.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr5_-_66776095 | 0.24 |
ENSMUST00000162366.8
ENSMUST00000162994.8 ENSMUST00000159512.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr8_+_26091607 | 0.23 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr3_+_125197722 | 0.23 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr2_-_38955518 | 0.22 |
ENSMUST00000039165.15
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr16_+_95059121 | 0.21 |
ENSMUST00000113858.3
|
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr17_-_48189815 | 0.21 |
ENSMUST00000154108.2
|
Foxp4
|
forkhead box P4 |
| chr1_-_155108455 | 0.21 |
ENSMUST00000035914.5
|
BC034090
|
cDNA sequence BC034090 |
| chr14_-_79461316 | 0.19 |
ENSMUST00000040802.5
|
Zfp957
|
zinc finger protein 957 |
| chr19_-_53577499 | 0.19 |
ENSMUST00000095978.5
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr6_-_124208815 | 0.18 |
ENSMUST00000233936.2
ENSMUST00000100968.4 |
Vmn2r27
|
vomeronasal 2, receptor27 |
| chr7_-_119461027 | 0.18 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr1_-_28819331 | 0.16 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr7_+_17799849 | 0.15 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr5_+_123648523 | 0.15 |
ENSMUST00000031384.6
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr3_-_129834788 | 0.14 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr19_-_11833365 | 0.13 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chr8_-_107064615 | 0.12 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr16_+_88966321 | 0.09 |
ENSMUST00000089098.2
|
Gm7735
|
predicted gene 7735 |
| chr9_-_110571645 | 0.09 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr9_+_56344700 | 0.08 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr17_+_35345292 | 0.07 |
ENSMUST00000061859.7
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr5_+_115573055 | 0.06 |
ENSMUST00000136586.6
|
Msi1
|
musashi RNA-binding protein 1 |
| chr13_+_16189041 | 0.05 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr1_+_167445815 | 0.05 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr8_+_26092252 | 0.05 |
ENSMUST00000136107.9
ENSMUST00000146919.8 ENSMUST00000139966.8 ENSMUST00000142395.8 ENSMUST00000143445.2 |
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr19_-_46033353 | 0.05 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr6_-_148732893 | 0.05 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr2_-_38955452 | 0.05 |
ENSMUST00000112850.9
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr9_+_111011388 | 0.04 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr3_+_63148887 | 0.04 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr2_-_88577230 | 0.03 |
ENSMUST00000099814.2
|
Olfr1198
|
olfactory receptor 1198 |
| chr3_-_95902949 | 0.03 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.7 | 5.2 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.5 | 12.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 9.4 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 1.2 | 32.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 1.0 | 3.0 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.9 | 8.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.8 | 4.0 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.8 | 8.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.7 | 4.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 2.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.6 | 1.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.6 | 13.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.5 | 2.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.4 | 3.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.4 | 1.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.4 | 3.4 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.4 | 1.4 | GO:2000293 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.3 | 4.3 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 2.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 1.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 1.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 2.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 3.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 1.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 2.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 0.5 | GO:0006404 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.7 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 0.9 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 0.8 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 0.8 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.2 | 3.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 3.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.7 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.8 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.6 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 7.5 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 1.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 3.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 2.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 2.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 2.2 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.5 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 4.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 3.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 2.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 3.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 5.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 3.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 2.8 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 32.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.7 | 3.9 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.4 | 4.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 9.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 2.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.3 | 4.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 0.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 3.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 12.4 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 0.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 2.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 3.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 6.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 7.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 7.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 4.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 12.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 4.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 8.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0030127 | ER to Golgi transport vesicle membrane(GO:0012507) COPII vesicle coat(GO:0030127) |
| 0.0 | 1.7 | GO:0043679 | axon terminus(GO:0043679) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 32.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 1.5 | 8.8 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.2 | 13.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 1.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.6 | 7.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.5 | 2.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.4 | 9.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 4.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 2.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 3.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 4.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 0.7 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.2 | 4.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 14.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 2.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 3.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 5.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 5.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 3.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 4.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 3.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 5.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 4.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 13.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 3.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |