Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
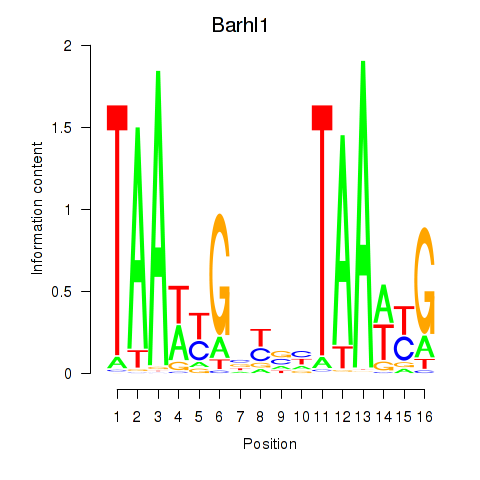
Results for Barhl1
Z-value: 1.10
Transcription factors associated with Barhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl1
|
ENSMUSG00000026805.15 | Barhl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl1 | mm39_v1_chr2_-_28806639_28806680 | -0.01 | 9.6e-01 | Click! |
Activity profile of Barhl1 motif
Sorted Z-values of Barhl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_75254989 | 9.89 |
ENSMUST00000039534.11
|
Resp18
|
regulated endocrine-specific protein 18 |
| chr6_-_41613322 | 5.96 |
ENSMUST00000031902.7
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr10_+_101994841 | 5.87 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr16_+_34815177 | 5.60 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chrX_+_162694397 | 5.20 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_+_27055245 | 4.76 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr8_+_31579499 | 4.65 |
ENSMUST00000036631.14
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr2_-_151474391 | 4.55 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr6_+_129568745 | 4.47 |
ENSMUST00000032268.14
ENSMUST00000112063.9 ENSMUST00000119520.8 |
Klrd1
|
killer cell lectin-like receptor, subfamily D, member 1 |
| chr3_-_146476331 | 4.41 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chrX_-_74621828 | 4.12 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr3_-_19319155 | 4.07 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr3_-_146475974 | 4.01 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr11_-_79418500 | 3.81 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr8_+_31579633 | 3.78 |
ENSMUST00000170204.8
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr11_+_116089678 | 3.63 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr6_-_130044234 | 3.63 |
ENSMUST00000119096.2
|
Klra4
|
killer cell lectin-like receptor, subfamily A, member 4 |
| chr2_+_83642910 | 3.54 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr10_+_101994719 | 3.45 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr16_+_92089268 | 3.42 |
ENSMUST00000047383.10
|
Kcne2
|
potassium voltage-gated channel, Isk-related subfamily, gene 2 |
| chr7_+_97492124 | 3.36 |
ENSMUST00000033040.12
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr5_+_149601688 | 3.20 |
ENSMUST00000100404.6
|
B3glct
|
beta-3-glucosyltransferase |
| chr2_-_104680104 | 3.14 |
ENSMUST00000028593.11
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr7_+_30463175 | 3.02 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr15_-_53765869 | 3.01 |
ENSMUST00000078673.14
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr5_+_14075281 | 3.00 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr14_-_9015639 | 2.71 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr3_+_75464837 | 2.62 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr3_-_75177378 | 2.61 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr15_-_58261093 | 2.51 |
ENSMUST00000227274.3
|
Anxa13
|
annexin A13 |
| chr11_+_61967821 | 2.49 |
ENSMUST00000092415.9
ENSMUST00000201015.4 ENSMUST00000202744.4 ENSMUST00000201723.4 ENSMUST00000202179.2 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr15_-_100579450 | 2.40 |
ENSMUST00000230740.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr6_-_129853617 | 2.36 |
ENSMUST00000014687.11
ENSMUST00000122219.2 |
Klra17
|
killer cell lectin-like receptor, subfamily A, member 17 |
| chr4_-_117013396 | 2.33 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chr15_-_100579813 | 2.27 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr10_+_58207229 | 2.27 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_130503041 | 2.21 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr17_-_25334879 | 2.18 |
ENSMUST00000024987.6
ENSMUST00000115181.9 |
Telo2
|
telomere maintenance 2 |
| chr17_-_37200927 | 2.14 |
ENSMUST00000087158.11
|
Trim40
|
tripartite motif-containing 40 |
| chr3_+_138019040 | 2.05 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr6_+_88879233 | 2.00 |
ENSMUST00000055022.15
ENSMUST00000204765.3 ENSMUST00000153874.8 |
Tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chrX_+_108240356 | 1.99 |
ENSMUST00000139259.2
ENSMUST00000060013.4 |
Gm6377
|
predicted gene 6377 |
| chr10_-_5019044 | 1.94 |
ENSMUST00000095899.5
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr2_-_73284262 | 1.90 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr17_+_7292967 | 1.83 |
ENSMUST00000097422.6
|
Gm1604b
|
predicted gene 1604b |
| chr3_+_115801869 | 1.80 |
ENSMUST00000106502.2
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr1_-_131441962 | 1.77 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr8_+_72635863 | 1.74 |
ENSMUST00000188685.7
|
Zfp709
|
zinc finger protein 709 |
| chr3_+_37402795 | 1.73 |
ENSMUST00000200585.5
ENSMUST00000038885.10 |
Fgf2
|
fibroblast growth factor 2 |
| chr9_+_19533374 | 1.65 |
ENSMUST00000213725.2
ENSMUST00000208694.2 |
Zfp317
|
zinc finger protein 317 |
| chr11_+_96209093 | 1.62 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr6_+_18170781 | 1.59 |
ENSMUST00000115406.2
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr4_+_112089442 | 1.58 |
ENSMUST00000038455.12
ENSMUST00000170945.2 |
Skint3
|
selection and upkeep of intraepithelial T cells 3 |
| chr2_+_122256913 | 1.58 |
ENSMUST00000110525.8
|
Slc28a2
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 |
| chr15_+_25752963 | 1.53 |
ENSMUST00000022882.12
ENSMUST00000135173.8 |
Myo10
|
myosin X |
| chr19_-_34143437 | 1.46 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr11_+_22921991 | 1.45 |
ENSMUST00000049506.8
|
Zrsr1
|
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 |
| chr11_+_54486925 | 1.42 |
ENSMUST00000218995.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr1_+_115612458 | 1.41 |
ENSMUST00000043725.9
|
Cntnap5a
|
contactin associated protein-like 5A |
| chr6_+_18170686 | 1.35 |
ENSMUST00000045706.12
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr15_-_43733389 | 1.28 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr5_+_90638580 | 1.23 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr9_+_19533591 | 1.22 |
ENSMUST00000215372.2
|
Zfp317
|
zinc finger protein 317 |
| chr14_+_25980463 | 1.21 |
ENSMUST00000173155.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr18_-_44308126 | 1.19 |
ENSMUST00000066328.5
|
Spinkl
|
serine protease inhibitor, Kazal type-like |
| chr6_+_123100382 | 1.18 |
ENSMUST00000032248.8
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr16_+_44687460 | 1.14 |
ENSMUST00000102805.4
|
Cd200r2
|
Cd200 receptor 2 |
| chr6_+_18170757 | 1.09 |
ENSMUST00000129452.8
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr3_-_49711706 | 1.07 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr2_-_45000389 | 1.06 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_41241049 | 1.06 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr5_-_23988551 | 1.05 |
ENSMUST00000148618.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr11_+_78717398 | 1.04 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr14_+_64097739 | 1.03 |
ENSMUST00000022528.6
|
Pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr6_-_130314465 | 1.01 |
ENSMUST00000088017.5
ENSMUST00000111998.9 |
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3 |
| chr10_+_128583734 | 0.99 |
ENSMUST00000163377.10
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr6_-_130170075 | 0.99 |
ENSMUST00000112032.8
ENSMUST00000071554.3 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr6_-_130003917 | 0.97 |
ENSMUST00000074056.3
|
Klra6
|
killer cell lectin-like receptor, subfamily A, member 6 |
| chr18_-_42395207 | 0.97 |
ENSMUST00000097590.5
|
Lars
|
leucyl-tRNA synthetase |
| chr17_+_37710293 | 0.94 |
ENSMUST00000216844.2
ENSMUST00000215974.2 ENSMUST00000215894.2 |
Olfr107
|
olfactory receptor 107 |
| chr16_-_32688640 | 0.92 |
ENSMUST00000089684.10
ENSMUST00000040986.15 ENSMUST00000115105.9 |
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr3_-_49711765 | 0.89 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr14_-_34225281 | 0.86 |
ENSMUST00000171343.9
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr18_+_37488174 | 0.86 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr18_+_44334331 | 0.85 |
ENSMUST00000097586.3
|
Gm10542
|
predicted gene 10542 |
| chr14_-_9015757 | 0.84 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr5_-_23988696 | 0.82 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr15_+_102240134 | 0.78 |
ENSMUST00000113682.9
ENSMUST00000001331.13 ENSMUST00000171244.2 |
Myg1
|
melanocyte proliferating gene 1 |
| chr6_+_42885812 | 0.70 |
ENSMUST00000216408.2
|
Olfr447
|
olfactory receptor 447 |
| chr17_-_32491339 | 0.69 |
ENSMUST00000237008.2
|
Brd4
|
bromodomain containing 4 |
| chr10_+_69623262 | 0.68 |
ENSMUST00000183240.2
|
Ank3
|
ankyrin 3, epithelial |
| chr4_+_98284005 | 0.66 |
ENSMUST00000238306.2
ENSMUST00000041284.10 |
Patj
|
PATJ, crumbs cell polarity complex component |
| chr3_+_32490300 | 0.66 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr3_-_19319123 | 0.65 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr2_-_45001141 | 0.63 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_86944911 | 0.59 |
ENSMUST00000216088.3
|
Olfr259
|
olfactory receptor 259 |
| chr2_-_111965322 | 0.56 |
ENSMUST00000213696.2
|
Olfr1316
|
olfactory receptor 1316 |
| chr2_-_79959178 | 0.54 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_-_41179167 | 0.52 |
ENSMUST00000043814.5
|
Fig4
|
FIG4 phosphoinositide 5-phosphatase |
| chr19_-_11816583 | 0.51 |
ENSMUST00000214887.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr9_+_37765565 | 0.50 |
ENSMUST00000213956.2
|
Olfr877
|
olfactory receptor 877 |
| chr14_-_26256025 | 0.49 |
ENSMUST00000139075.8
ENSMUST00000102956.8 |
Slmap
|
sarcolemma associated protein |
| chr15_+_28203872 | 0.47 |
ENSMUST00000067048.8
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr7_-_103113358 | 0.44 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr16_-_59459745 | 0.42 |
ENSMUST00000099646.10
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr11_+_101473062 | 0.41 |
ENSMUST00000039581.14
ENSMUST00000100403.9 ENSMUST00000107194.8 ENSMUST00000128614.2 |
Tmem106a
|
transmembrane protein 106A |
| chr14_+_53093071 | 0.38 |
ENSMUST00000181038.3
ENSMUST00000187138.2 |
Trav14d-1
|
T cell receptor alpha variable 14D-1 |
| chr9_+_38179147 | 0.33 |
ENSMUST00000212156.3
|
Olfr895
|
olfactory receptor 895 |
| chr3_+_37402495 | 0.33 |
ENSMUST00000138563.9
|
Fgf2
|
fibroblast growth factor 2 |
| chr18_-_44329234 | 0.31 |
ENSMUST00000239421.2
ENSMUST00000097587.5 |
Spink11
|
serine peptidase inhibitor, Kazal type 11 |
| chr1_-_63253702 | 0.28 |
ENSMUST00000050536.14
|
Gpr1
|
G protein-coupled receptor 1 |
| chr12_-_54842488 | 0.23 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chr2_+_86338805 | 0.22 |
ENSMUST00000076263.2
|
Olfr1076
|
olfactory receptor 1076 |
| chr14_+_53599724 | 0.19 |
ENSMUST00000196105.2
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chrX_-_163033364 | 0.18 |
ENSMUST00000112263.2
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr7_-_103214484 | 0.17 |
ENSMUST00000106886.2
|
Olfr616
|
olfactory receptor 616 |
| chr18_-_42395131 | 0.13 |
ENSMUST00000236102.2
|
Lars
|
leucyl-tRNA synthetase |
| chr11_-_69127848 | 0.11 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr16_-_16681839 | 0.10 |
ENSMUST00000100136.4
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr2_-_88860295 | 0.10 |
ENSMUST00000216592.2
ENSMUST00000213669.2 |
Olfr1217
|
olfactory receptor 1217 |
| chr10_-_126866682 | 0.08 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr18_+_46730765 | 0.08 |
ENSMUST00000238168.2
ENSMUST00000078079.11 ENSMUST00000168382.2 ENSMUST00000235849.2 ENSMUST00000235973.2 ENSMUST00000235455.2 ENSMUST00000237478.2 |
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr9_+_38312994 | 0.05 |
ENSMUST00000214648.3
ENSMUST00000056364.3 |
Olfr147
|
olfactory receptor 147 |
| chr8_-_85567256 | 0.05 |
ENSMUST00000003911.13
ENSMUST00000109761.9 ENSMUST00000128035.2 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr10_+_129539079 | 0.01 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 1.7 | 8.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.6 | 4.8 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 1.1 | 5.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 3.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.8 | 3.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.7 | 2.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.7 | 4.7 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.6 | 8.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.5 | 1.6 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.4 | 6.0 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 1.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.4 | 3.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.4 | 1.8 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.3 | 1.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 1.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 1.2 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.3 | 0.9 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.3 | 1.9 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 2.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 0.7 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 2.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 1.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 2.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 3.6 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 0.9 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 1.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 2.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 5.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 1.2 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 3.0 | GO:2000249 | negative regulation of cell-matrix adhesion(GO:0001953) regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 2.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 4.5 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 4.5 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 1.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 5.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 5.0 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 8.0 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.4 | 9.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.2 | 3.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.6 | 4.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.5 | 8.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 1.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 2.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 2.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 2.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 3.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 3.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 5.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 4.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 5.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 3.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 10.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.4 | 8.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.3 | 4.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 1.1 | 4.5 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.8 | 9.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.8 | 8.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.6 | 5.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 1.8 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.4 | 1.6 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.4 | 3.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.4 | 1.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.3 | 1.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 2.5 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.2 | 0.7 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.2 | 3.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 4.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 4.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 2.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 5.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 3.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 5.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.7 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 7.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 6.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 5.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 9.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 6.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 9.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 9.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 8.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 4.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 3.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 5.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 4.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 5.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |