Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
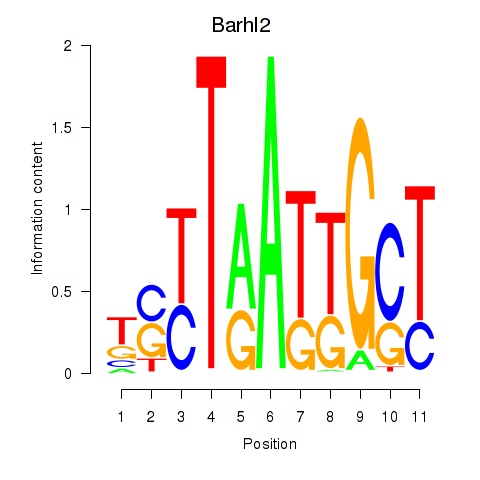
Results for Barhl2
Z-value: 1.01
Transcription factors associated with Barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl2
|
ENSMUSG00000034384.12 | Barhl2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl2 | mm39_v1_chr5_-_106606032_106606032 | -0.60 | 1.9e-08 | Click! |
Activity profile of Barhl2 motif
Sorted Z-values of Barhl2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34506744 | 9.50 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr5_+_110987839 | 9.15 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr7_+_24069680 | 9.15 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr2_-_103114105 | 8.99 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr11_-_100098333 | 7.20 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr17_+_34457868 | 7.03 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr11_+_62711057 | 6.87 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr7_+_100145192 | 6.46 |
ENSMUST00000133044.3
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_+_62711295 | 6.41 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr5_-_110987604 | 6.20 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr3_-_49711765 | 5.82 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr3_-_49711706 | 5.63 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr5_-_110987441 | 5.57 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr5_-_134975773 | 5.44 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr10_-_44024843 | 5.15 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr5_+_110988095 | 4.89 |
ENSMUST00000198373.2
|
Chek2
|
checkpoint kinase 2 |
| chr19_-_6947112 | 4.74 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chr17_+_34544632 | 4.72 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr17_+_35481702 | 4.69 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr7_+_127845984 | 4.63 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr6_-_136758716 | 4.50 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr7_+_43086432 | 4.45 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr7_+_116103599 | 4.33 |
ENSMUST00000032895.15
|
Nucb2
|
nucleobindin 2 |
| chr7_+_127846121 | 4.28 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr6_+_125529911 | 4.26 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr7_+_116103643 | 4.20 |
ENSMUST00000183175.8
|
Nucb2
|
nucleobindin 2 |
| chr3_-_95158457 | 4.00 |
ENSMUST00000125515.3
ENSMUST00000107195.9 |
Bnipl
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr8_+_57774010 | 3.94 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr4_-_127224591 | 3.93 |
ENSMUST00000046532.4
|
Gjb3
|
gap junction protein, beta 3 |
| chr2_+_143757193 | 3.80 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr10_+_58230203 | 3.67 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_+_57964921 | 3.56 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr10_+_39488930 | 3.52 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr7_+_43086554 | 3.49 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr8_+_57964956 | 3.35 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr3_-_95158360 | 3.31 |
ENSMUST00000098871.11
ENSMUST00000137250.9 |
Bnipl
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr2_-_126975804 | 3.27 |
ENSMUST00000110387.4
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr5_+_92535705 | 3.14 |
ENSMUST00000138687.2
ENSMUST00000124509.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr2_+_118644717 | 3.08 |
ENSMUST00000028803.14
ENSMUST00000126045.8 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr7_+_28580847 | 3.04 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr8_-_62576140 | 3.03 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr2_-_147887810 | 3.00 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr11_+_95733489 | 2.89 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr12_-_104439589 | 2.85 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr10_+_58230183 | 2.82 |
ENSMUST00000020077.11
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr13_+_51799268 | 2.78 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr2_+_125876566 | 2.72 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr16_-_36648586 | 2.72 |
ENSMUST00000023537.6
ENSMUST00000114829.9 |
Eaf2
|
ELL associated factor 2 |
| chr16_+_38182569 | 2.67 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr18_+_4993795 | 2.65 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr7_+_126810780 | 2.60 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr1_+_171265103 | 2.58 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr7_+_79309938 | 2.57 |
ENSMUST00000035977.9
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr11_-_120464062 | 2.53 |
ENSMUST00000026122.11
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr7_+_110368037 | 2.48 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr11_+_96209093 | 2.44 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr11_+_116324913 | 2.37 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr5_+_33815466 | 2.28 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr7_+_113113061 | 2.24 |
ENSMUST00000129087.8
ENSMUST00000067929.15 ENSMUST00000164745.8 ENSMUST00000136158.8 |
Far1
|
fatty acyl CoA reductase 1 |
| chr9_+_30853837 | 2.23 |
ENSMUST00000068135.13
|
Adamts8
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
| chr7_+_113113037 | 2.23 |
ENSMUST00000033018.15
|
Far1
|
fatty acyl CoA reductase 1 |
| chr11_-_120463667 | 2.23 |
ENSMUST00000168360.2
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr2_+_57887896 | 2.22 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr15_+_98972850 | 2.18 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein |
| chr6_-_101354858 | 2.07 |
ENSMUST00000075994.11
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr17_-_25946370 | 2.02 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr5_-_120726721 | 1.99 |
ENSMUST00000046426.10
|
Tpcn1
|
two pore channel 1 |
| chr2_+_125876883 | 1.97 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr6_-_8259098 | 1.91 |
ENSMUST00000012627.5
|
Rpa3
|
replication protein A3 |
| chr9_-_110453427 | 1.87 |
ENSMUST00000196876.2
ENSMUST00000035069.14 |
Nradd
|
neurotrophin receptor associated death domain |
| chr2_-_181101158 | 1.86 |
ENSMUST00000155535.2
ENSMUST00000029106.13 ENSMUST00000087409.10 |
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr5_-_65855511 | 1.83 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr5_+_92535532 | 1.83 |
ENSMUST00000145072.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr12_+_24701273 | 1.81 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr15_-_101482320 | 1.78 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr11_-_40646090 | 1.76 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr3_-_129518723 | 1.74 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr9_+_61279640 | 1.73 |
ENSMUST00000178113.8
ENSMUST00000159386.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr15_+_101371353 | 1.69 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr16_+_10652910 | 1.66 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr4_-_96673423 | 1.66 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr6_-_34294377 | 1.64 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr7_-_48497771 | 1.57 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr14_+_53698556 | 1.48 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr7_-_19449319 | 1.45 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr2_+_30282266 | 1.44 |
ENSMUST00000028209.15
|
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr6_-_136899167 | 1.42 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr3_+_95496239 | 1.40 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr11_+_95733109 | 1.40 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr5_+_135197228 | 1.34 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr3_+_95496270 | 1.30 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr2_-_151822114 | 1.29 |
ENSMUST00000062047.6
|
Fam110a
|
family with sequence similarity 110, member A |
| chr18_+_34869412 | 1.28 |
ENSMUST00000105038.3
|
Gm3550
|
predicted gene 3550 |
| chr17_+_35439810 | 1.28 |
ENSMUST00000118793.2
|
Gm16181
|
predicted gene 16181 |
| chr7_-_126975199 | 1.26 |
ENSMUST00000067425.6
|
Zfp747
|
zinc finger protein 747 |
| chr11_+_60308077 | 1.23 |
ENSMUST00000070681.7
|
Gid4
|
GID complex subunit 4, VID24 homolog |
| chr2_+_30282414 | 1.20 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr8_+_18896267 | 1.14 |
ENSMUST00000149565.8
ENSMUST00000033847.5 |
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr18_+_61178211 | 1.12 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr4_-_58553553 | 1.06 |
ENSMUST00000107575.9
ENSMUST00000107574.8 ENSMUST00000147354.8 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr7_+_43093507 | 1.05 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr7_+_101619019 | 1.04 |
ENSMUST00000084852.13
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr3_-_154034271 | 1.04 |
ENSMUST00000204403.2
|
Lhx8
|
LIM homeobox protein 8 |
| chr17_-_47813201 | 1.02 |
ENSMUST00000233174.2
ENSMUST00000233121.2 ENSMUST00000067103.4 |
Taf8
|
TATA-box binding protein associated factor 8 |
| chr6_-_5496261 | 1.00 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr5_-_115236354 | 1.00 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr6_+_70821481 | 0.99 |
ENSMUST00000034093.15
ENSMUST00000162950.2 |
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr11_-_116743898 | 0.99 |
ENSMUST00000190993.7
ENSMUST00000092404.13 |
Srsf2
|
serine and arginine-rich splicing factor 2 |
| chr19_+_12839106 | 0.98 |
ENSMUST00000059675.4
|
Olfr1444
|
olfactory receptor 1444 |
| chr11_+_3599183 | 0.97 |
ENSMUST00000096441.5
|
Morc2a
|
microrchidia 2A |
| chr2_+_9887427 | 0.97 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr5_+_88523967 | 0.96 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr10_-_99595498 | 0.95 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr4_+_132495636 | 0.94 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr18_-_78249612 | 0.92 |
ENSMUST00000163367.3
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr14_+_52892115 | 0.91 |
ENSMUST00000198019.2
|
Trav7-1
|
T cell receptor alpha variable 7-1 |
| chr2_-_45002902 | 0.91 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr7_-_98790275 | 0.90 |
ENSMUST00000037968.10
|
Uvrag
|
UV radiation resistance associated gene |
| chr5_-_28672091 | 0.89 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr2_+_3514071 | 0.86 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr9_+_56979307 | 0.82 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr4_-_43710231 | 0.82 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr4_-_9643636 | 0.82 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr14_+_53007210 | 0.81 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr9_-_13738204 | 0.81 |
ENSMUST00000147115.8
|
Cep57
|
centrosomal protein 57 |
| chr2_+_31530049 | 0.81 |
ENSMUST00000113470.3
|
Prdm12
|
PR domain containing 12 |
| chr5_+_135197137 | 0.79 |
ENSMUST00000031692.12
|
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr7_-_126975169 | 0.78 |
ENSMUST00000205832.2
|
Zfp747
|
zinc finger protein 747 |
| chr4_-_120427449 | 0.77 |
ENSMUST00000030381.8
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chr14_-_30850795 | 0.77 |
ENSMUST00000049732.11
ENSMUST00000090205.5 ENSMUST00000064032.10 |
Smim4
|
small integral membrane protein 4 |
| chrX_-_167970196 | 0.75 |
ENSMUST00000066112.12
ENSMUST00000112118.8 ENSMUST00000112120.8 ENSMUST00000112119.8 |
Amelx
|
amelogenin, X-linked |
| chr11_-_60307847 | 0.74 |
ENSMUST00000108721.9
ENSMUST00000239016.2 |
Atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr8_-_84831391 | 0.72 |
ENSMUST00000041367.9
ENSMUST00000210279.2 |
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr1_+_165616250 | 0.72 |
ENSMUST00000161971.8
ENSMUST00000187313.7 ENSMUST00000178336.8 ENSMUST00000005907.12 ENSMUST00000027849.11 |
Cd247
|
CD247 antigen |
| chr7_+_127440924 | 0.71 |
ENSMUST00000033075.14
|
Stx4a
|
syntaxin 4A (placental) |
| chr12_+_100745314 | 0.69 |
ENSMUST00000069782.11
|
Dglucy
|
D-glutamate cyclase |
| chr2_+_118731860 | 0.68 |
ENSMUST00000036578.7
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr7_+_101619327 | 0.65 |
ENSMUST00000210475.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr11_-_69791756 | 0.65 |
ENSMUST00000018714.13
ENSMUST00000128046.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr7_-_12063408 | 0.64 |
ENSMUST00000226701.2
|
Vmn1r83
|
vomeronasal 1 receptor 83 |
| chr15_-_34356567 | 0.64 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr14_-_21764638 | 0.64 |
ENSMUST00000073870.7
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr10_-_23968192 | 0.63 |
ENSMUST00000092654.4
|
Taar8b
|
trace amine-associated receptor 8B |
| chr11_-_58443063 | 0.63 |
ENSMUST00000073933.2
|
Olfr328
|
olfactory receptor 328 |
| chr7_+_126359869 | 0.60 |
ENSMUST00000206272.2
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chrX_-_156198282 | 0.59 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr11_-_69791712 | 0.58 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr16_-_63684477 | 0.58 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr18_+_42644552 | 0.55 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr11_+_49327451 | 0.54 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr14_+_53328803 | 0.53 |
ENSMUST00000103609.2
|
Trav7n-4
|
T cell receptor alpha variable 7N-4 |
| chr18_-_84104507 | 0.53 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr7_+_3632982 | 0.53 |
ENSMUST00000179769.8
ENSMUST00000008517.13 |
Prpf31
|
pre-mRNA processing factor 31 |
| chr13_+_23966524 | 0.53 |
ENSMUST00000074067.4
|
Trim38
|
tripartite motif-containing 38 |
| chr16_-_63684425 | 0.51 |
ENSMUST00000232049.2
|
Epha3
|
Eph receptor A3 |
| chr19_-_13828056 | 0.51 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr6_-_146403638 | 0.49 |
ENSMUST00000079573.13
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr7_+_127441119 | 0.49 |
ENSMUST00000121705.8
|
Stx4a
|
syntaxin 4A (placental) |
| chr11_-_99494134 | 0.49 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr11_-_69791774 | 0.47 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr1_+_173983199 | 0.46 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr18_-_84104574 | 0.43 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr19_-_13827773 | 0.41 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr19_-_4861536 | 0.41 |
ENSMUST00000179909.8
ENSMUST00000172000.3 ENSMUST00000180008.2 ENSMUST00000113793.4 ENSMUST00000006625.8 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr7_+_113113292 | 0.40 |
ENSMUST00000122890.2
|
Far1
|
fatty acyl CoA reductase 1 |
| chr4_-_43000450 | 0.40 |
ENSMUST00000030164.8
|
Vcp
|
valosin containing protein |
| chr13_+_118851214 | 0.39 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr2_-_88531671 | 0.38 |
ENSMUST00000104892.2
|
Olfr1196
|
olfactory receptor 1196 |
| chr1_+_74401267 | 0.38 |
ENSMUST00000097697.8
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr4_+_19818718 | 0.36 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr6_+_8259379 | 0.31 |
ENSMUST00000162034.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr9_-_105009023 | 0.31 |
ENSMUST00000035179.9
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr18_+_61820982 | 0.31 |
ENSMUST00000025471.4
|
Il17b
|
interleukin 17B |
| chr7_+_24967094 | 0.30 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chr14_-_30850881 | 0.28 |
ENSMUST00000203261.3
|
Smim4
|
small integral membrane protein 4 |
| chr5_+_20112771 | 0.26 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr19_-_11838430 | 0.25 |
ENSMUST00000214796.2
|
Olfr1418
|
olfactory receptor 1418 |
| chrX_+_138464065 | 0.25 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr8_-_25528972 | 0.23 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr9_-_105008972 | 0.22 |
ENSMUST00000186925.2
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr7_-_12002196 | 0.21 |
ENSMUST00000227973.2
ENSMUST00000228764.2 ENSMUST00000228482.2 ENSMUST00000227080.2 |
Vmn1r81
|
vomeronasal 1 receptor 81 |
| chr3_-_69034425 | 0.21 |
ENSMUST00000194558.6
ENSMUST00000029353.9 |
Kpna4
|
karyopherin (importin) alpha 4 |
| chr7_+_126359763 | 0.19 |
ENSMUST00000091328.4
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr15_-_100322089 | 0.19 |
ENSMUST00000154331.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr10_+_18345706 | 0.17 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr2_-_84545504 | 0.16 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr15_+_66542598 | 0.16 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr11_+_112673041 | 0.15 |
ENSMUST00000000579.3
|
Sox9
|
SRY (sex determining region Y)-box 9 |
| chr3_-_54621774 | 0.15 |
ENSMUST00000178832.2
|
Gm21958
|
predicted gene, 21958 |
| chr14_-_99337137 | 0.15 |
ENSMUST00000042471.11
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr5_+_146885450 | 0.15 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr19_-_37340010 | 0.14 |
ENSMUST00000131070.3
|
Ide
|
insulin degrading enzyme |
| chrX_+_168662592 | 0.13 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr3_+_96537484 | 0.11 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr1_+_153767478 | 0.10 |
ENSMUST00000050660.6
|
Teddm1a
|
transmembrane epididymal protein 1A |
| chr3_+_96537235 | 0.09 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr1_+_105918383 | 0.08 |
ENSMUST00000119166.8
|
Zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr11_-_100504159 | 0.07 |
ENSMUST00000146840.3
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr9_+_39657545 | 0.04 |
ENSMUST00000213358.2
|
Olfr967
|
olfactory receptor 967 |
| chr10_-_107321938 | 0.04 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.0 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 2.8 | 8.5 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 2.3 | 7.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.7 | 5.1 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.5 | 11.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.0 | 7.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.0 | 13.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.0 | 3.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.9 | 4.7 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.9 | 8.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.8 | 3.3 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.8 | 4.9 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.7 | 6.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.7 | 4.7 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.7 | 7.3 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.6 | 3.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.6 | 1.9 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.6 | 6.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 | 9.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.6 | 3.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.5 | 1.6 | GO:0090420 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.5 | 2.7 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.5 | 4.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.5 | 1.6 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.5 | 9.5 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.4 | 1.7 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.4 | 1.2 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.4 | 1.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.4 | 2.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.4 | 1.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 2.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 1.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.3 | 2.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 6.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 | 1.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 11.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 0.8 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.3 | 1.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 2.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 1.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 2.5 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.2 | 2.7 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 2.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.8 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.2 | 0.8 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 5.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 0.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 3.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 2.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 0.9 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 2.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.7 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 3.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.8 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.4 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.1 | 2.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.5 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 1.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 2.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 4.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 4.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.0 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.8 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 1.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 8.2 | GO:0001890 | placenta development(GO:0001890) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 2.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.7 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 1.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 2.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.0 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.7 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 1.9 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 2.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 5.9 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 2.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 16.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.0 | 4.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.9 | 2.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.7 | 3.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.5 | 13.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 4.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.7 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.3 | 1.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 1.1 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 1.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 10.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 4.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 5.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 4.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.2 | 21.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 3.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 4.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 3.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 7.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 15.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 5.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 5.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 8.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 7.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 8.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.1 | 6.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.0 | 7.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.0 | 13.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 1.0 | 9.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.0 | 4.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.8 | 4.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.7 | 5.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.7 | 6.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.7 | 2.6 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.5 | 3.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.5 | 2.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 2.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 1.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.4 | 14.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.4 | 5.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 1.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 7.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.7 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 1.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 2.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 4.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.9 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.2 | 1.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 9.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 2.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 4.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 0.5 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.2 | 4.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 0.5 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.2 | 8.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 3.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 2.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.9 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 1.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 4.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 2.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 3.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 13.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 1.1 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 14.0 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 4.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 9.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 8.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 11.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 3.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 14.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 6.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 4.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 4.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 13.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 6.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 8.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 5.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 4.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 4.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 3.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 7.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 9.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 2.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |