Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Batf
Z-value: 0.60
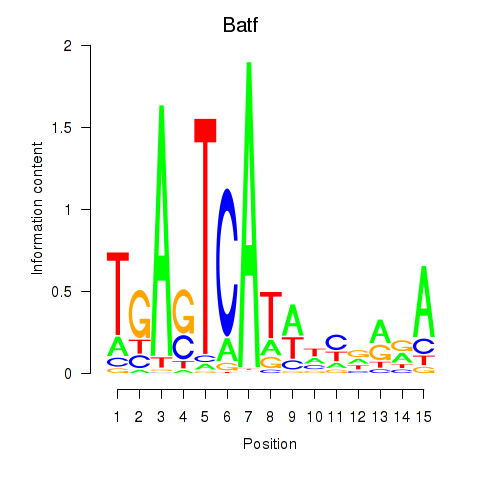
Transcription factors associated with Batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf
|
ENSMUSG00000034266.7 | Batf |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf | mm39_v1_chr12_+_85733415_85733478 | -0.12 | 3.1e-01 | Click! |
Activity profile of Batf motif
Sorted Z-values of Batf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_31383976 | 6.74 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr7_-_44861235 | 3.78 |
ENSMUST00000210741.2
ENSMUST00000209466.2 |
Dkkl1
|
dickkopf-like 1 |
| chr1_+_134313676 | 3.45 |
ENSMUST00000162187.2
|
Mgat4f
|
MGAT4 family, member F |
| chr5_+_92524813 | 3.04 |
ENSMUST00000120781.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr2_-_155356716 | 2.95 |
ENSMUST00000029131.11
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr2_-_126625343 | 2.93 |
ENSMUST00000028842.9
ENSMUST00000130356.3 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr11_-_97913420 | 2.62 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chrY_-_1286623 | 2.44 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr6_-_40917431 | 2.43 |
ENSMUST00000122181.8
ENSMUST00000031935.10 |
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr7_-_133966588 | 2.43 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr5_+_137759934 | 2.26 |
ENSMUST00000110983.3
ENSMUST00000031738.5 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr7_+_30487322 | 2.19 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr5_+_31274046 | 2.16 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr9_+_78522783 | 2.11 |
ENSMUST00000093812.5
|
Cd109
|
CD109 antigen |
| chr5_-_114229413 | 1.89 |
ENSMUST00000150106.2
|
Svop
|
SV2 related protein |
| chrX_+_132751729 | 1.84 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr18_-_43071297 | 1.79 |
ENSMUST00000153737.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_177635997 | 1.59 |
ENSMUST00000194528.3
|
Catspere1
|
cation channel sperm associated auxiliary subunit epsilon 1 |
| chr3_-_10248979 | 1.40 |
ENSMUST00000029034.9
|
Pmp2
|
peripheral myelin protein 2 |
| chr1_+_182392577 | 1.40 |
ENSMUST00000048941.14
|
Capn8
|
calpain 8 |
| chr4_+_109092459 | 1.40 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr11_-_29775649 | 1.34 |
ENSMUST00000104962.2
|
Fem1al
|
fem-1 homolog A like |
| chr7_-_144305705 | 1.31 |
ENSMUST00000155175.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chrX_-_152838035 | 1.28 |
ENSMUST00000144098.2
|
Samt1c
|
spermatogenesis associated multipass transmembrane protein 1c |
| chr4_+_109092610 | 1.24 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr16_-_59421342 | 1.23 |
ENSMUST00000172910.3
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chrX_-_152816823 | 1.22 |
ENSMUST00000177994.8
|
Samt1b
|
spermatogenesis associated multipass transmembrane protein 1b |
| chrX_-_152795636 | 1.20 |
ENSMUST00000178342.8
|
Samt1
|
spermatogenesis associated multipass transmembrane protein 1 |
| chr10_-_117074501 | 1.19 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chrX_-_152903681 | 1.17 |
ENSMUST00000112565.2
|
Samt1d
|
spermatogenesis associated multipass transmembrane protein 1d |
| chr3_-_145813802 | 1.16 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3 |
| chr3_+_64884839 | 1.16 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_+_182392559 | 1.14 |
ENSMUST00000168514.7
|
Capn8
|
calpain 8 |
| chr19_+_4644425 | 1.10 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr1_+_107456731 | 1.06 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr10_+_128542120 | 1.06 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chr18_+_57605525 | 1.03 |
ENSMUST00000209786.2
|
Ctxn3
|
cortexin 3 |
| chr10_-_83369994 | 1.02 |
ENSMUST00000020497.14
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr13_-_55979191 | 1.00 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr5_+_17779273 | 1.00 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chrX_+_51880056 | 0.97 |
ENSMUST00000101588.2
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr18_+_82928959 | 0.97 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr17_-_54153367 | 0.96 |
ENSMUST00000023886.7
|
Sult1c2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr8_+_41276027 | 0.90 |
ENSMUST00000066814.7
|
Adam39
|
a disintegrin and metallopeptidase domain 39 |
| chr10_+_87926932 | 0.85 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr13_-_32960379 | 0.85 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr2_-_74409225 | 0.84 |
ENSMUST00000134168.2
ENSMUST00000111993.9 ENSMUST00000064503.13 |
Lnpk
|
lunapark, ER junction formation factor |
| chr18_-_35631914 | 0.81 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr1_+_165130192 | 0.79 |
ENSMUST00000111450.3
|
Gpr161
|
G protein-coupled receptor 161 |
| chr11_-_3864664 | 0.76 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr9_+_113641615 | 0.75 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr7_+_98092628 | 0.75 |
ENSMUST00000098274.5
|
Gucy2d
|
guanylate cyclase 2d |
| chr5_-_44256528 | 0.74 |
ENSMUST00000196178.2
ENSMUST00000197750.5 |
Prom1
|
prominin 1 |
| chr4_-_155953849 | 0.68 |
ENSMUST00000030950.8
|
Cptp
|
ceramide-1-phosphate transfer protein |
| chr15_+_102922247 | 0.68 |
ENSMUST00000001709.3
|
Hoxc5
|
homeobox C5 |
| chr5_-_86666408 | 0.67 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr2_-_111320501 | 0.63 |
ENSMUST00000099616.2
|
Olfr1290
|
olfactory receptor 1290 |
| chr15_-_8739893 | 0.60 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr3_+_59637203 | 0.59 |
ENSMUST00000168156.3
|
Aadacl2fm2
|
AADACL2 family member 2 |
| chr14_+_55459462 | 0.58 |
ENSMUST00000022820.12
|
Dhrs2
|
dehydrogenase/reductase member 2 |
| chr4_+_58285957 | 0.56 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr2_-_74409723 | 0.56 |
ENSMUST00000130586.8
|
Lnpk
|
lunapark, ER junction formation factor |
| chr10_+_79304565 | 0.54 |
ENSMUST00000233065.2
ENSMUST00000167976.4 |
Vmn2r83
|
vomeronasal 2, receptor 83 |
| chr5_+_16996808 | 0.51 |
ENSMUST00000211738.2
|
Gm28710
|
predicted gene 28710 |
| chr13_+_27380623 | 0.50 |
ENSMUST00000018066.13
|
Prl3c1
|
prolactin family 3, subfamily c, member 1 |
| chr3_+_103034843 | 0.50 |
ENSMUST00000172288.3
|
Dennd2c
|
DENN/MADD domain containing 2C |
| chr5_+_30218112 | 0.48 |
ENSMUST00000026845.12
ENSMUST00000199183.5 ENSMUST00000195978.5 |
Il6
|
interleukin 6 |
| chr7_-_142215595 | 0.47 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chrY_-_13465579 | 0.46 |
ENSMUST00000178428.2
|
Gm21866
|
predicted gene, 21866 |
| chr9_+_108782646 | 0.46 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr3_+_84081411 | 0.45 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr17_+_25105617 | 0.44 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr11_+_96941420 | 0.43 |
ENSMUST00000090020.13
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr4_-_117232650 | 0.43 |
ENSMUST00000094853.9
|
Rnf220
|
ring finger protein 220 |
| chr9_+_108782664 | 0.41 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr18_+_37100672 | 0.40 |
ENSMUST00000193777.6
ENSMUST00000193389.2 |
Pcdha6
|
protocadherin alpha 6 |
| chr6_+_89691185 | 0.38 |
ENSMUST00000075158.2
|
Vmn1r40
|
vomeronasal 1 receptor 40 |
| chr7_-_133384449 | 0.32 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr7_+_126376353 | 0.32 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr2_-_52448552 | 0.30 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr4_+_109092829 | 0.30 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr2_-_157179344 | 0.29 |
ENSMUST00000109536.8
|
Ghrh
|
growth hormone releasing hormone |
| chr9_-_39914761 | 0.29 |
ENSMUST00000215523.2
|
Olfr979
|
olfactory receptor 979 |
| chr6_-_136812423 | 0.28 |
ENSMUST00000068293.4
ENSMUST00000111894.2 |
Smco3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr11_+_96941637 | 0.27 |
ENSMUST00000168565.2
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr3_-_79439181 | 0.26 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr4_-_59783780 | 0.25 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chrY_+_41361843 | 0.25 |
ENSMUST00000178559.2
|
Gm21800
|
predicted gene, 21800 |
| chr2_+_88285211 | 0.22 |
ENSMUST00000219871.2
ENSMUST00000213115.3 |
Olfr1183
|
olfactory receptor 1183 |
| chr13_-_65051763 | 0.22 |
ENSMUST00000091554.6
|
Cntnap3
|
contactin associated protein-like 3 |
| chr11_-_78950698 | 0.21 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr9_+_37766116 | 0.21 |
ENSMUST00000086063.4
|
Olfr877
|
olfactory receptor 877 |
| chr17_+_48080113 | 0.20 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr7_-_142215027 | 0.19 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr18_+_82928782 | 0.18 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516 |
| chr5_-_6926523 | 0.18 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr7_-_102408573 | 0.17 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr3_+_93107517 | 0.17 |
ENSMUST00000098884.4
|
Flg2
|
filaggrin family member 2 |
| chr13_-_8921732 | 0.17 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr3_-_122828592 | 0.16 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr3_-_63391300 | 0.16 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr6_-_136812377 | 0.14 |
ENSMUST00000203468.2
|
Smco3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr4_-_127251917 | 0.14 |
ENSMUST00000046498.3
|
Gjb5
|
gap junction protein, beta 5 |
| chr7_-_79244677 | 0.14 |
ENSMUST00000172788.3
|
Rhcg
|
Rhesus blood group-associated C glycoprotein |
| chr7_-_85895409 | 0.14 |
ENSMUST00000165771.2
ENSMUST00000233075.2 ENSMUST00000233317.2 ENSMUST00000233312.2 ENSMUST00000233928.2 ENSMUST00000232799.2 ENSMUST00000233744.2 |
Vmn2r76
|
vomeronasal 2, receptor 76 |
| chr7_+_126376319 | 0.13 |
ENSMUST00000132643.2
|
Ypel3
|
yippee like 3 |
| chr2_-_36995660 | 0.13 |
ENSMUST00000100144.2
|
Olfr362
|
olfactory receptor 362 |
| chr17_-_25105277 | 0.12 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chr14_-_32907023 | 0.12 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr8_-_27903965 | 0.12 |
ENSMUST00000033882.10
|
Chrna6
|
cholinergic receptor, nicotinic, alpha polypeptide 6 |
| chr14_+_26789345 | 0.11 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
| chr7_+_115692530 | 0.10 |
ENSMUST00000032899.12
ENSMUST00000106608.8 ENSMUST00000106607.2 |
1110004F10Rik
|
RIKEN cDNA 1110004F10 gene |
| chr7_+_108312527 | 0.10 |
ENSMUST00000209620.4
ENSMUST00000074730.4 |
Olfr512
|
olfactory receptor 512 |
| chr2_-_134396268 | 0.09 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr7_+_101714943 | 0.08 |
ENSMUST00000094130.4
ENSMUST00000084843.10 |
Xndc1
Xntrpc
|
Xrcc1 N-terminal domain containing 1 Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
| chr3_-_37286714 | 0.08 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr6_-_144993451 | 0.08 |
ENSMUST00000123930.8
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr18_+_42408418 | 0.08 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr2_-_91274967 | 0.07 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr11_+_81936531 | 0.07 |
ENSMUST00000021011.3
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr14_-_32907446 | 0.06 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr7_-_12829100 | 0.05 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr4_-_118795809 | 0.04 |
ENSMUST00000215312.3
|
Olfr1328
|
olfactory receptor 1328 |
| chr18_+_82929037 | 0.02 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr18_+_82929451 | 0.02 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr6_-_122259768 | 0.01 |
ENSMUST00000032207.9
|
Klrg1
|
killer cell lectin-like receptor subfamily G, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 0.8 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.2 | 1.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 1.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 3.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 2.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 2.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 1.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.6 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 1.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 2.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 1.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 3.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.2 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 8.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 2.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.7 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 3.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.3 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 1.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 2.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 2.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 5.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 3.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 3.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 2.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 1.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.8 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 5.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 2.9 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |